Staging data (cancer)¶
The 100kGP cancer_staging_consolidated LabKey table compiles in one place all the staging information available in the Genomics England research environment. It contains the subset of participants from the cancer programme who have successfully passed through the Genomics England interpretation pipeline (and are available in the cancer_analysis LabKey table) for whom at least one piece of staging information is found.
Please note that cancer_staging_consolidated contains no new staging information, i.e. no information that is not already in other LabKey tables.
Description¶
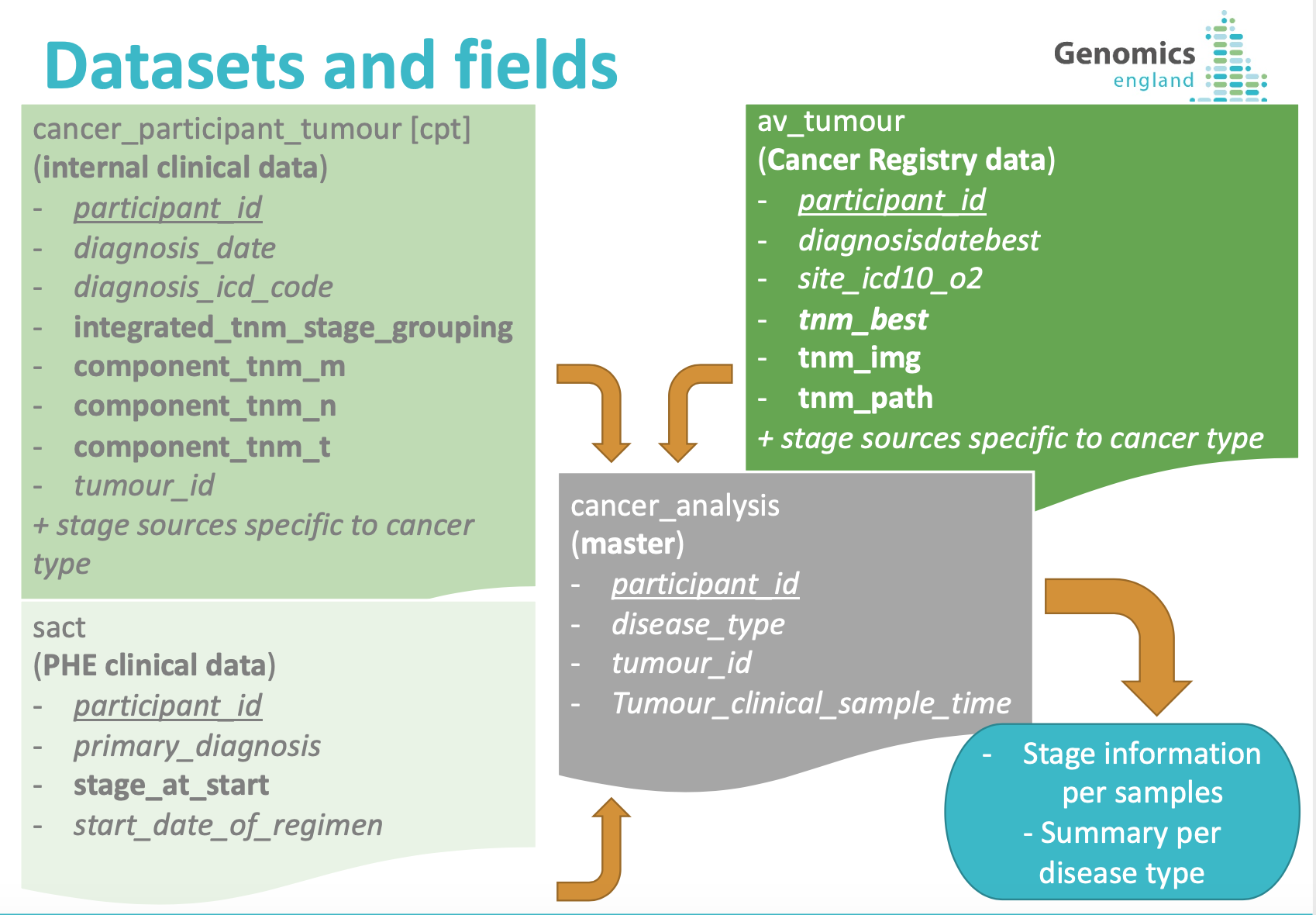
Staging information is located in the Research Environment in the following three datasets:
cancer_participant_tumour(Genomic England primary clinical data)av_tumour(secondary clinical data from NCRAS)sact(secondary clinical data from NHSE)
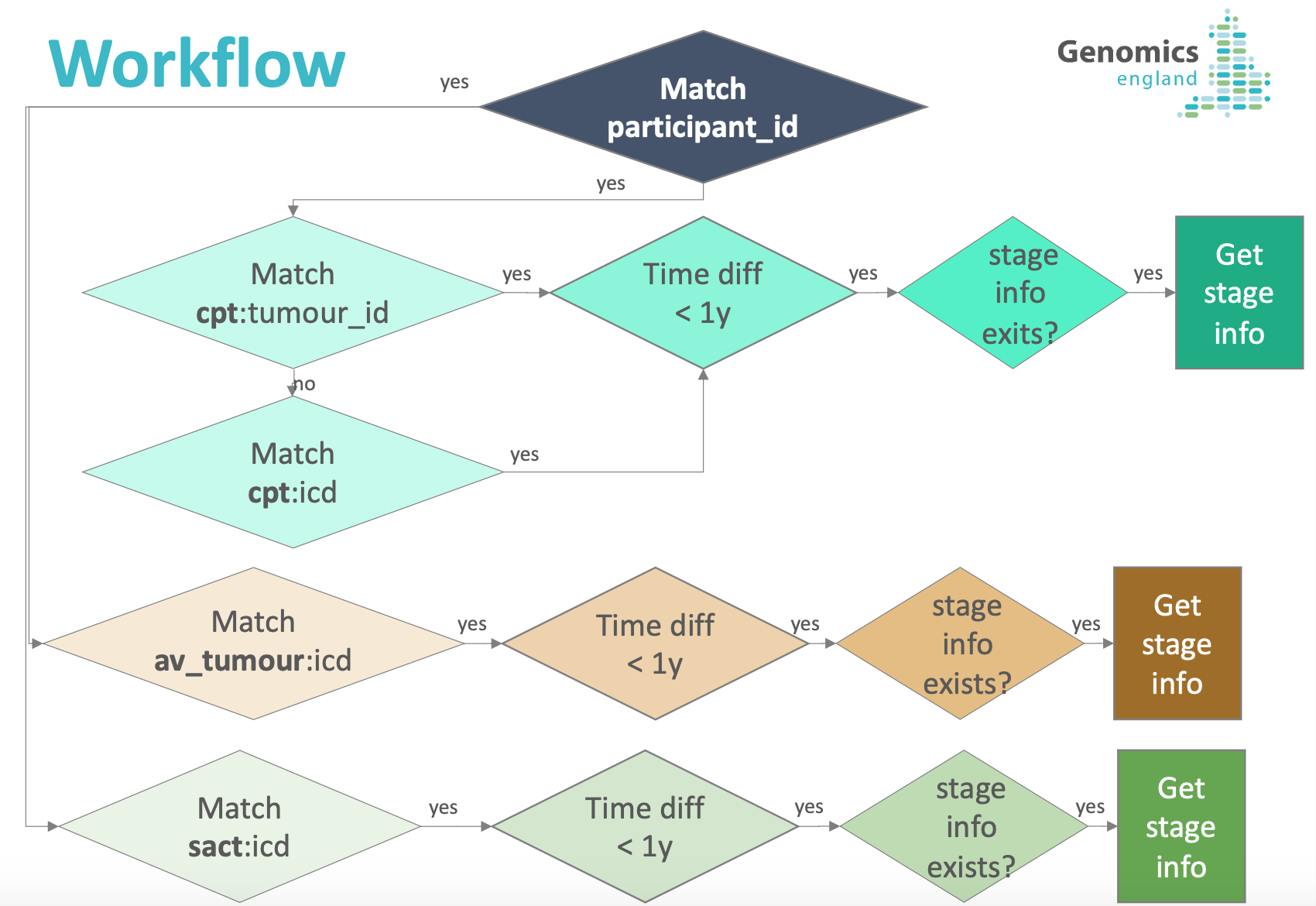
These datasets have different levels of completion for the staging information. In addition, all tables on LabKey are linked via participant_id, which in the case of cancer staging data is not sufficient, since one participant can have multiple tumours and stage will evolve with time. In order to make staging information easily accessible, we have put together, in a single table, the staging information found on the datasets above for each tumour sample in cancer_analysis.
Tumour_id made it possible to link samples with our primary clinical data; however, not all samples had a tumour_id available. In these cases, as well as for the secondary clinical data, samples have been linked using a dictionary that correlates ICD-10 codes found in the clinical data and disease_type of cancer_analysis. The dictionary was create internally and validated by one of our pathologists.
Finally, we only include staging information in the cancer_staging_consolidated table when the available clinical stage information has been collected no more than one year (12 months) from the date when the tumour sample was collected. If you would like to use a smaller window, you can do so by filtering on column "interval_min" of cancer_staging_consolidated table (please note that the interval_min is counted in days). If for a tumour sample there are multiple staging information available within the one year window, only one entry per source dataset (cancer_participant_tumour, av_tumour, sact) will be included: the staging information that was obtained closer to the date when the tumour sample was collected. For sact data, we link samples using participant_id and disease_type and use the starting date of regimen; if there is a match (via disease_type), we ensure that the starting date of regimen and the date the tumour sample was taken are no more than one year apart.
Staging and grading information available¶
| Staging type | Column header | Definition | Cancer type | Further information |
|---|---|---|---|---|
| TNM | integrated_tnm_stage_grouping |
The overall integrated TNM stage grouping indicates the tumour stage after treatment and/or after all available evidence has been collected. | Any | Link |
component_tnm_t |
Tumour stage, if integrated TNM not supplied. This is the UICC code which classifies the size and extent of the primary tumour after treatment and/or after all available evidence has been collected. | Any | Link | |
component_tnm_n |
Nodes stage, if integrated TNM not supplied. This is the UICC code which classifies the absence or presence and extent of regional lymph node metastases after treatment and/or after all available evidence has been collected | Any | Link | |
component_tnm_m |
Metastasis stage, if integrated TNM not supplied. This is the UICC code which classifies the absence or presence of distant metastases after treatment and/or after all available evidence has been collected. | Any | Link | |
t_best |
The best tumour stage out of t_path and t_img, based on the shortest time-lapse after diagnosis. |
Any | Link | |
n_best |
The best nodes stage out of n_path and n_img, based on the shortest time-lapse after diagnosis. |
Any | Link | |
m_best |
The best metastasis stage out of m_path and m_img, based on the shortest time-lapse after diagnosis. |
Any | Link | |
t_path |
Tumour stage, determined from pathology data | Any | Link | |
n_path |
Nodes stage, determined from pathology data | Any | Link | |
m_path |
Metastasis stage, determined from pathology data | Any | Link | |
t_img |
Tumour stage, determined from image data | Any | Link | |
n_img |
Nodes stage, determined from image data | Any | Link | |
m_img |
Metastasis stage, determined from image data | Any | Link | |
| AJCC | ajcc_stage |
American Joint Committee on Cancer staging of tumour at diagnosis. | Skin cancer | |
| FIGO | figo |
Fédération Internationale de Gynécologie et d’Obstétrique staging | ovarian, endometrial, cervical, vaginal and vulval cancer | Link |
final_figo_stage |
FIGO stage following surgery for uterine and vulval malignancies and for ovarian malignancies undergoing primary surgery. For ovarian malignancies planned to undergo neoadjuvant chemotherapy and for cases of cervical cancer (which is staged clinically), the final FIGO stage is determined at the time of review of clinical findings, imaging, cytology and biopsy histology. | ovarian, endometrial, cervical, vaginal and vulval cancer | Link | |
| Dukes | dukes |
Dukes' stage | Bowel cancer | Link |
modified_dukes_stage |
Dukes' stage of disease at diagnosis (based on pathological evidence but upgraded to Dukes D if clinical evidence of metastasis) Dukes D should be recorded if metastatic spread is identified either in the preoperative staging process, e.g. on CT scanning, MRI, USS, chest x-ray or at the time of operation. It is accepted that a small number of D cases are cured by further treatment such as liver resection, but for COSD metastatic spread distant from the primary should always be recorded as D. | Bowel cancer | Link | |
| Stage | stage_best |
Best ‘registry’ stage at diagnosis of the tumour | All | |
stage_best_system |
System used to record best registry stage at diagnosis | All | ||
stage_path |
Stage based on pathology | All | ||
stage_img |
Stage based on imaging | All | ||
| Gleason | gleason_primary |
Gleason primary pattern | Prostate cancer | Link |
gleason_combined |
Combined Gleason primary and secondary scores | Prostate cancer | Link | |
| Grade | grade |
Grade of Differentiation, how abnormal the cancer cells are | Any | Link |
| Oestrogen receptor status | er_status |
Low levels of oestrogen receptor | Breast cancer | Link |
| Progesterone receptor status | pr_status |
Low levels of progesterone receptor | Breast cancer | Link |
| HER2 status | her2_status |
Elevated levels of human epidermal growth factor 2 | Breast cancer | Link |
| Nottingham Prognostic Index | npi |
a calculation of the probability of success of surgery for breast cancer | Breast cancer | Link |
Workflow¶
Location¶
This information can be found in LabKey under a tabled called cancer_staging_consolidated under the Bioinformatics tab. The cancer_staging_consolidated table connects to other tables in LabKey via the participant_id. In addition, tumour identifiers from different sources, i.e. tumour_id (Genomics England), av_tumour_pseudo_id (NCRAS) and sact_tumour_pseudo_id (NHSE) are given to identify the specific tumour.
Table schema¶
The cancer_staging_consolidated table contains the following entries:
from cancer_analysis:
- participant_id
- tumour_sample_platekey
- tumour_id
- disease_type
- tumour_type
- tumour_clinical_sample_time
from cancer_participant_tumour:
- diagnosis_date
- diagnosis_icd_code
- integrated_tnm_stage_grouping
- component_tnm_t
- component_tnm_n
- component_tnm_m
- ajcc_stage
- final_figo_stage
- modified_dukes_stage
from av_tumour:
- av_tumour_pseudo_id
- diagnosisdatebest
- site_icd10_o2
- stage_best
- t_best
- n_best
- m_best
- stage_best_system
- stage_path
- t_path
- n_path
- m_path
- stage_img
- t_img
- n_img
- m_img
- dukes
- figo
- gleason_primary
- gleason_combined
- grade
- behaviour_coded_desc
- histology_coded_desc
- er_status
- pr_status
- her2_status
- npi
from sact:
- sact_tumour_pseudo_id
- primary_diagnosis
- start_date_of_regimen
- stage_at_start
calculated:
- interval_min
Cancer staging statistics¶
The statistics for the cancer_staging_consolidated table for Cancer staging V8 data release can be found here: Cancer staging V8 Statistics (28-11-2019)
The statistics for the cancer_staging_consolidated table for Cancer staging V9 data release can be found here: Cancer staging V9 Statistics (02-04-2020)
The statistics for the cancer_staging_consolidated table for Cancer staging V10 data release can be found here: Cancer staging V10 Statistics (03-09-2020)
The statistics for the cancer_staging_consolidated table for Cancer staging V11 data release can be found here: Cancer staging V11 Statistics (17-12-2020)
The statistics for the cancer_staging_consolidated table for Cancer staging V12 data release can be found here: Cancer staging V12 Statistics (06-05-2021)
The statistics for the cancer_staging_consolidated table for Cancer staging V13 data release can be found here: Cancer staging V13 Statistics (30-09-2021)
The statistics for the cancer_staging_consolidated table for Cancer staging V14 data release can be found here: Cancer staging V14 Statistics (27-01-2022)
The statistics for the cancer_staging_consolidated table for Cancer staging V15 data release can be found here: Cancer staging V15 Statistics (26-05-2022)
The statistics for the cancer_staging_consolidated table for Cancer staging V16 data release can be found here: Cancer staging V16 Statistics (13-10-2022)
The statistics for the cancer_staging_consolidated table for Cancer staging V17 data release can be found here: Cancer staging V17 Statistics (30-03-2023)
The statistics for the cancer_staging_consolidated table for Cancer staging V18 data release can be found here: Cancer staging V18 Statistics (21-12-2023)
The statistics for the cancer_staging_consolidated table for Cancer staging V19 data release can be found here: Cancer staging V19 Statistics (31-10-2024)
Feedback¶
This table has been included for the first time in data release 8. If you have suggestions about this table or would like to request edits that would be useful for your analyses, please let us know by contacting us via the Genomics England Service Desk.