Data in the Research Environment¶
The main types of data in the Research Environment are:
- Clinical and phenotype data data for each participant
- Genomic data for each participant from our sequencing providers
- Genomic and associated data from the Genomics England bioinformatic pipelines
- Publicly available genomic datasets and cohorts
- Research Community provided data
At Genomics England all identifiable information is stripped out and associated to the patient's participant_id so that all patients' data can be linked to their clinical and genomic data.
Data sources¶
There are two sources of data available in the RE:
- The main programme or 100,000 Genomes Project data
- The NHS Genomic Medicine Service
Main programme or 100,000 Genomes Project¶
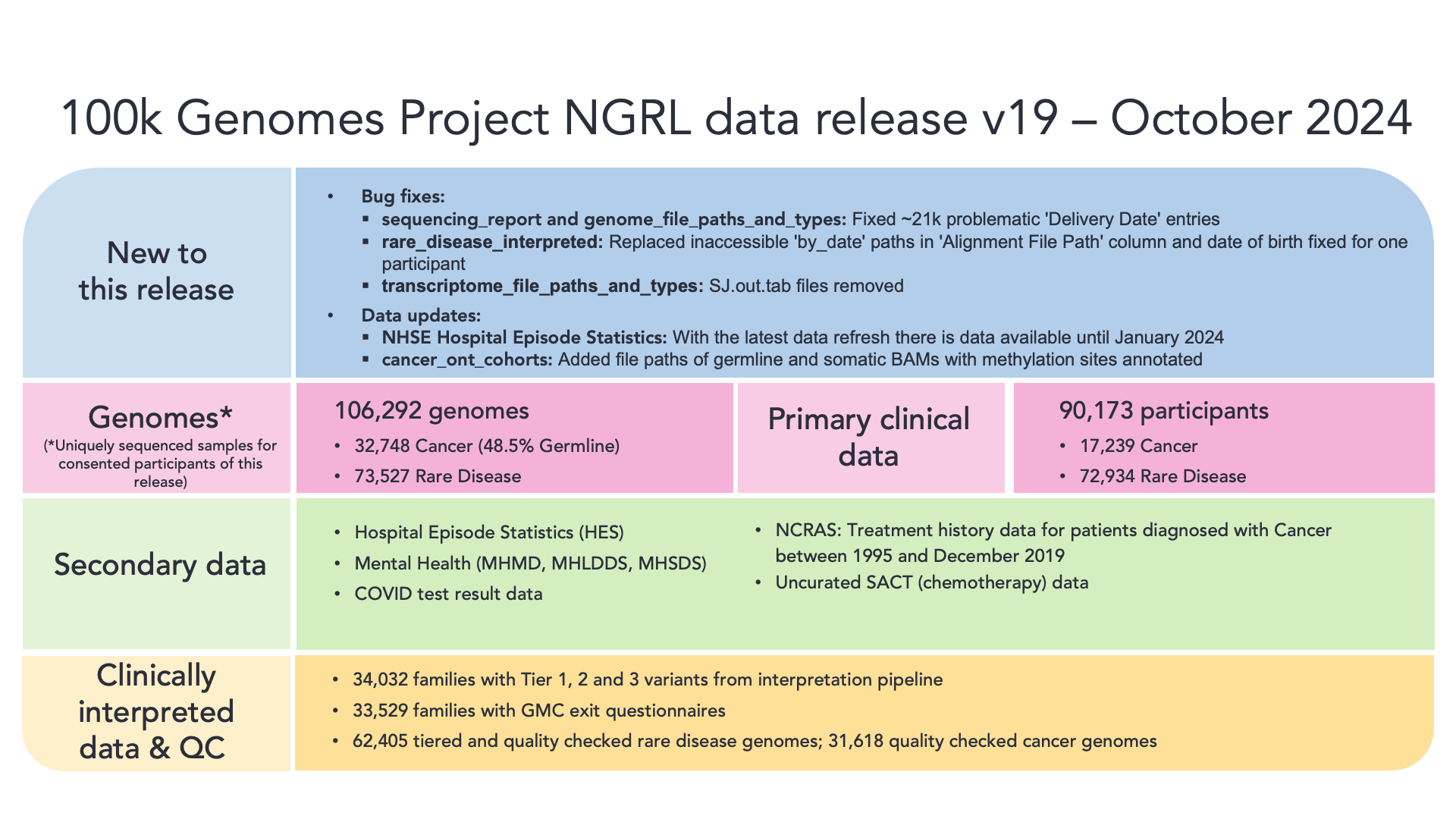
The 100,000 Genomes Project was a British initiative to sequence and study the role our genes play in health and disease. This involved sequencing the genomes of 85,000 cancer and rare disease participants. Data from these participants are available as the main programme in the Genomics England RE. The medical histories of these participants are continuously updated, but there will be no new genomes added.
You can find details of the current release, and an overview below:
NHS GMS programme¶
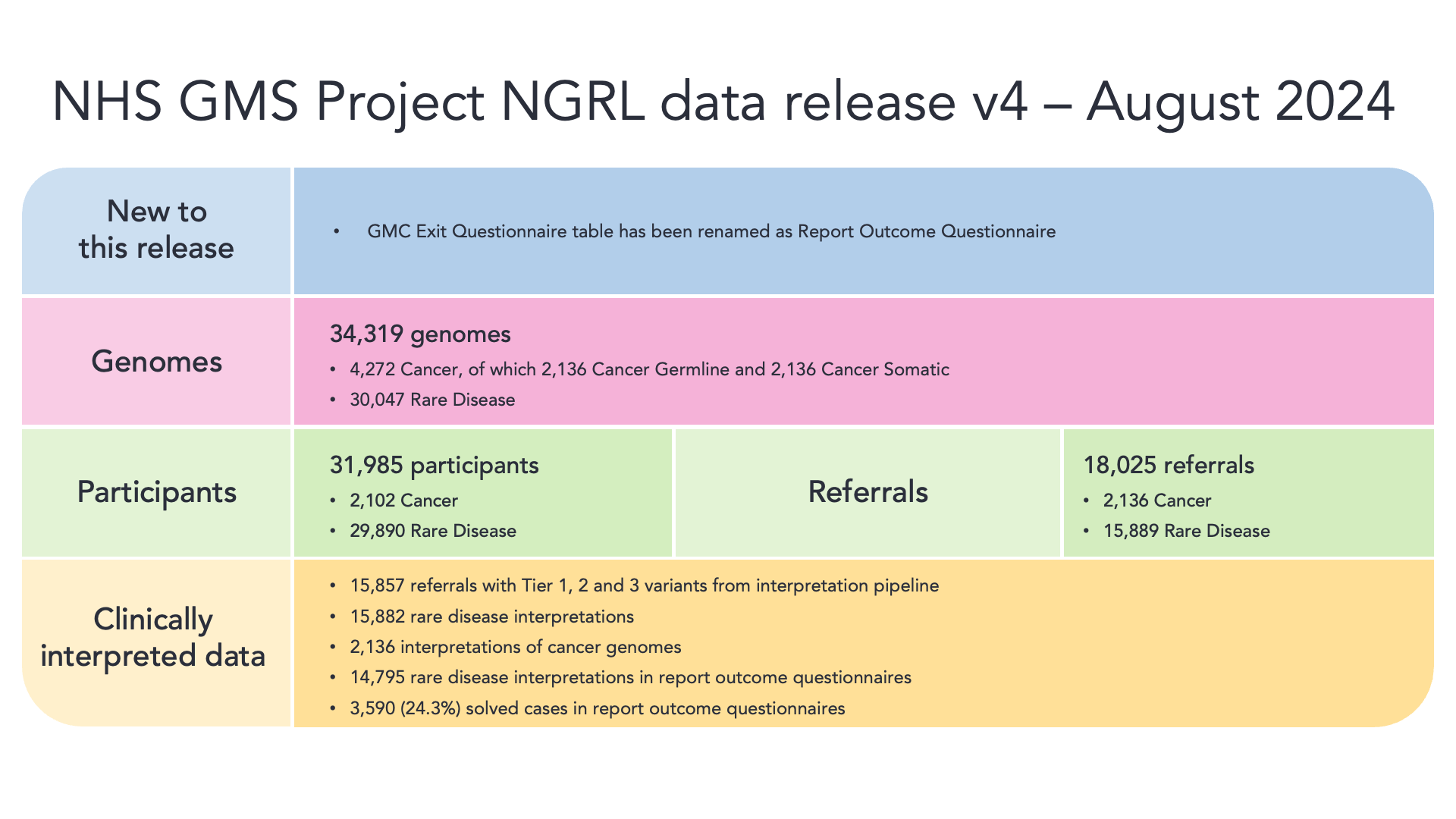
Genomics England now provides a continual sequencing service for the NHS Genomic Medicine Service, sequencing genomes of rare disease and cancer participants where these are required in clinical practice in the NHS. We will continuously receive new participants in the NHS GMS programme.
You can find details of the current release, and an overview below:
Clinical and phenotype data¶
The clinical and phenotype data are stored in a data management application called LabKey which is accessible from the Research Environment Desktop. Clinical and phenotype data are sourced from the GMCs according to set data models that specify the variables and matching data types. These include hospital episode statistics tables from NHS England and disease registration data from NDRS. Not all variables are compulsory and some will contain personal identifiable data, so are not present in the de-identified data within the Research Environment. Participant phenotypes such as age, sex, ethnicity, pedigree, recruited disease, associated HPO terms, and tumour categories can be analysed by using the LabKey application.
These tables also contain the results of some bioinformatic analysis, as described under Genomics England data.
There are datasets for the 100kGP project and from the NHS GMS data ingestion.
Genomic data¶
The genomic data delivered by our sequencing provider are provided as a genome delivery which includes BAMs and VCFs for each participant. The genomic data are accessed through the file system where each genome delivery represents a unique sequenced genome. All available genomes are in the genomes folder, organised by date of delivery.
Genomics England data¶
A subset of the genomic and associated data from the Genomics England bioinformatic pipelines are also provided through the file system. These data include files which were necessary for genome interpretation; such as the joint-called by family VCFs, tiered variants, aggregated variant calls, and internal allele frequency files. You can access some these files by navigating to the gel_data_resources folder. Some of the data is included in the LabKey tables.
Publicly available data¶
The publicly available genomic datasets and cohorts includes datasets such as: 1000 Genomes data, reference genomes for both GRCh37 and GRCh38 assemblies, BLAST databases, CADD databases and many others. These can be accessed through our file system by navigating to the Home icon on the Research Environment desktop and selecting the public_data_resources folder. You are able to request additional publicly available data at any time by contacting the Service Desk.
Research community provided data¶
Results of analyses by our collaborators, using Genomics England data, and made available through the RE.
Our future plans¶
You can keep up-to-date with our plans for data in the RE, give us feedback on those plans and make suggestions using our Productboard roadmap.
Video tutorial¶
Look to see how these files are organised in this video tutorial: