Variant Effect Predictor (VEP) container¶
VEP is a tool provided by Ensembl for functional annotation of variant consequences on genes. We currently provide versions 99, 105, 106, 109 and 111 of VEP as containers. These include the most used plugins.
Description¶
The Variant Effect Predictor (VEP) is a comprehensive annotation source created by the Ensembl project and relies on their database to provide annotation data for variants. To facilitate the use of this tool and conform to the need to ensure the security of the data that we host we have developed containerised versions of the tool which include the canonical plugins.
The container has been created from a Docker image converted to a Singularity file. The contents listed in the section below:
VEP v.111 release
The VEP 111 container includes:
* the [AlphaMissense plugin](https://github.com/Ensembl/VEP_plugins/blob/release/111/AlphaMissense.pm) has been included
* The project base image has been updated to Ubuntu 22.04
* the following plugins have been removed as they user Python 2.x which is no longer supported:
* FATHMM
* FATHMM_MKL
* PON_P2
We have had a change in the way we support the various versions of LOFTEE, this is due to the way the new base image functions. Effectively we need to release two separate containers for Build 37 and Build 38. We have made these containers available in our internal mirror (Artifactory):
For Build 37:
docker://docker-gel-research-containers.artifactory.aws.gel.ac/vep:v111_lofteeb37
For Build 38:
docker://docker-gel-research-containers.artifactory.aws.gel.ac/vep:v111_lofteeb38
Contents of the VEP containers
- VEP (https://www.ensembl.org/info/docs/tools/vep/index.html)
- Plugin list (for more information see http://www.ensembl.org/info/docs/tools/vep/script/vep_plugins.html):
- AlphaMissense plugin (v111 only)
- AncestralAllele
- Blosum62
- CADD
- Carol
- Condel
- Conservation
- dbNSFP
- dbscSNV
- DisGeNET
- Downstream
- Draw
- ExACpLI
- ExAC
- FATHMM_MKL (not v110)
- FATHMM (not v110)
- FlagLRG
- FunMotifs
- G2P
- GeneSplicer
- gnomADc
- GO
- HGVSIntronOffset
- LD
- LocalID
- LoFtool
- loftee-GRCh37
- loftee-GRCh38
- LOVD
- Mastermind
- MaxEntScan
- MPC
- MTR
- NearestExonJB
- NearestGene
- neXtProt
- Phenotypes
- plugin_config.txt
- PON_P2 (not v110)
- PostGAP
- ProteinSeqs
- ReferenceQuality
- REVEL
- SameCodon
- satMutMPRA
- SingleLetterAA
- SpliceAI
- SpliceRegion
- StructuralVariantOverlap
- SubsetVCF
- TSSDistance
Notes¶
The container included all plugins currently available within the Ensembl VEP project's GitHub page with the addition of LOFTEE. Other third party plugins may be made available on a case-by-case basis, however, due to the inherent limitations within VEP, the annotation of VCFs with these third party tools cannot be combined with the internal plugins in a single step.
It is important to note the following points:
- The container will overwrite any existing output files, please ensure that any prior work you have performed is backed up before launching a new annotation.
- The annotation jobs need to be submitted to the HPC either via an interactive job for a single file or submitted as a batch of jobs for a list of files, please consult this page for instructions on how to achieve this.
- Because of the way filesystems are mounted on the HPC you will need to mount (--bind) the full path to the locations for your input, output and other ancillary files please refer to the examples below to guide you.
Commercial usage
If you are a Discovery Forum member, there are some restrictions on the plugins and VEP options you are allowed to use.
Genomics England imposes no restrictions on access to, or use of, the data provided and the software used to analyse and present it.
Some of the data and software included in the distribution may be subject to third-party constraints. You are solely responsible for establishing the nature of and complying with any such restrictions.
Instructions¶
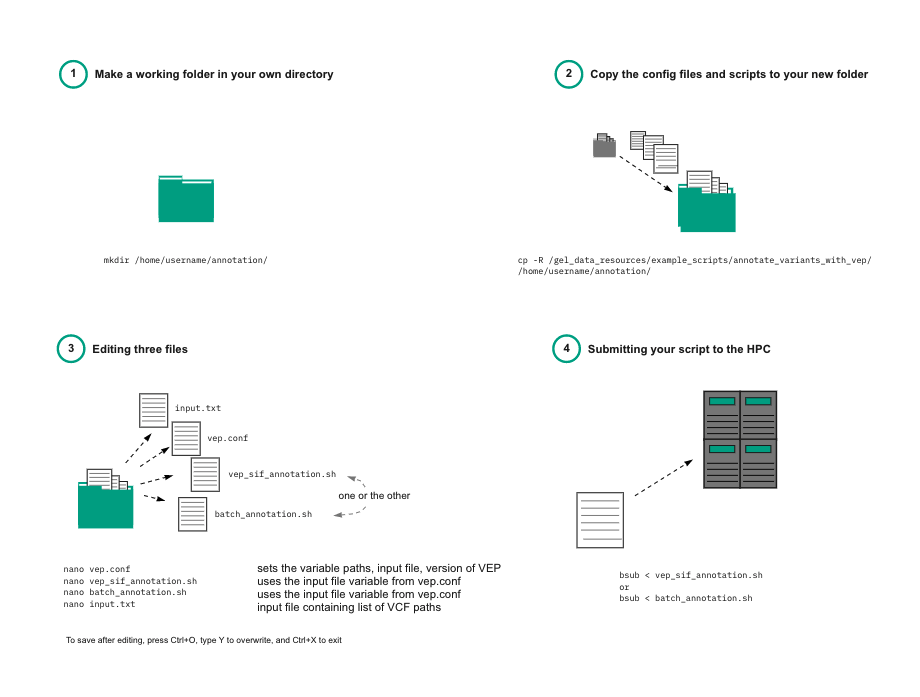
Using the container¶
For versions 99, 105, 106, 109, and 111 copy the folder to your working directory:
cp -R /gel_data_resources/example_scripts/annotate_variants_with_vep/<VERSION> /<PATH_TO_YOUR_WORKING_DIRECTORY>/
This will copy the submission scripts and configuration files for the selected version of the VEP container.
Create an input file list¶
If you are only annotating a single VCF file, then you only need to add this to your submission script. However, if you're annotating multiple VCFs, you need to create a file list.
You should use the latest version of LabKey to compile your list.
Example of an input file
Save the file to your working directory.
Edit the configuration file¶
Most of the necessary folder locations required for executing the container are already included in the config file vep_###.conf.
If you are annotating multiple VCFs, you will need to edit the INPUT_FILE variable with the name of your list of VCFs.
Edit the submission script¶
Edit the annotation script called vep_sif_annotation.sh. You will need to edit
* `#BSUB -q <queue_name>`: the queue that you want to submit the script to. If it is the first time you are trying it, we recommend the *short* queue (`#BSUB -q short`) as it will run for four hours at most.
* `#BSUB -P <your_project_code>`: your [project code](lsf_codes.md). For example `#BSUB -P re_gecip_neurology`.
* `input =`: the file you want to annotate.
* `output_file` in the VEP options: The full file path for the output.
A sample of VEP options and plugins have been included in the submission script. You can edit the VEP command to add or remove options and plugins as specified in the Ensembl VEP documentation.
Edit the annotation script called batch_annotation.sh.
* `#BSUB -q <queue_name>`: Specify the queue that you want to submit the script to. We recommend using the *medium* (24 hours max) or *long* (one week default but changeable) queues for this script (`#BSUB -q long`).
* `#BSUB -P <your_project_code>`: Specify your [project code](lsf_codes.md). For example `#BSUB -P re_gecip_neurology`.
* `output_file` in the VEP options: The full file path for the output.
A sample of VEP options and plugins have been included in the submission script. You can edit the VEP command to add or remove options and plugins as specified in the Ensembl VEP documentation.
Submit the script¶
Submit to the HPC using:
bsub < vep_sif_annotation.sh
Submit to the HPC using:
bsub < batch_annotation.sh
Inputs and outputs¶
| File or Folder | Description | Requires editing |
|---|---|---|
| vep_sif_annotation.sh | Single VCF file annotation | Yes, can be provided by you. Example only. |
| batch_annotation.sh | Batch VCF file annotation | Yes, can be provided by you. Example only. |
| vep.conf | Configuration file for VEP variables | Yes. Example only. |
| input.txt | List of vcf file paths. These will either be extracted from LabKey as part of your cohort building process or the /path/to/VCF/files if you are using VCF files custom called and aggregated VCFs. The process requires that there be one path per line. | Yes |