Cancer survival analysis¶
This scripts combines Genomics England secondary data to support survival analysis. This script is meant to be used as an guide on where to find relevant information, keeping in mind that each cohort should be further curated and that real-world evidence requires data cleaning and quality checks.
The code can be found here:
/gel_data_resources/example_scripts/survival_oncology
Please copy it into your dir and edit according to your needs.
Summary¶
The script will provide the basis for a cohort build, estimate time of survival or until last time seen, and combine with genetic data.
| Usage | Variables | Columns | Table | Comment |
|---|---|---|---|---|
| cohort building | patient id sample id |
participant_id tumour_sample_platekey |
cancer_analysis | This can be further subset. av_tumour is a great source of information for such. Please make use of the tutorial on cohort building to build you own custom cohorts. |
| survival time | death last seen date of diagnosis |
participant_id date_of_death/death_date |
||
arrivaldate/disdate/apptdate/ccdisdate diagnosis_date+diagnosis_icd_code/ diagnosisdatebest+site_icd10_o2 |
mortality/death_details ae/apc/op/cc cancer_participant_tumour |
Consider selecting only survival <5 years Note that diagnosis code is also select to confirm that the date of diagnosis corresponds to the correct disease |
||
| SNV status | mutations on a gene of interest | uses gene_centric-report, please read here on how to select your variants: Gene centric SNV report for cancer participants |
Missing diagnosis dates
For a subset of cases, the diagnosis date is missing. In these cases, the data is imputed.
Pre-requirements¶
Make sure you have LabKey API working on your machine. You also need to install some R dependencies. Some of these will only work on R v4.2.1 or later.
Getting started¶
Open a terminal and type:
install the dependencies:
dependencies <- c("tidyverse", "lubridate", "survminer", "survival", "Rlabkey", "gdata", "bit64")
install.packages(dependencies, lib = '~/path/to/personal/library/directory')
These libraries will have a number of their own dependencies so may take some time to be installed.
Input¶
You need to edit the gene of interest, in this case BRCA2.
Search for SNVdb to localise the relevant part of code, change the gene name for more information, such as selecting specific variants, look at: Gene centric SNV report for cancer participants
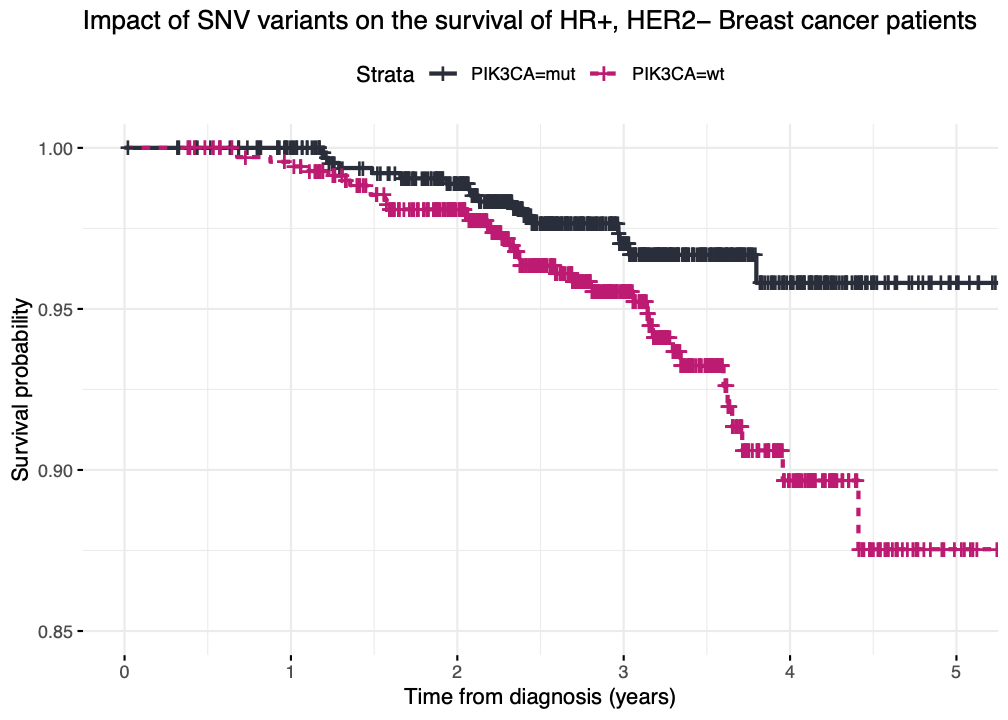
Output¶
A survival curve split for reference and alternative alleles of a certain gene.
All tables relevant to retrieve survival, including cleaning how to clean the data should be listed here.
Example of a plot created based on the script above, applying specific changes for the cohort of interest.