NHS genomic medicine service data release v3 (18/03/2024)¶
Purpose¶
This document provides a description of the NHS GMS data release v3 dated 18th March 2024.
Each progressive release incorporates new content, enhances existing content, and enables more effective use of the data.
This data are presented within the Genomics England Research Environment, accessed via the AWS virtual desktop interface and subject to all Genomics England data protection and privacy principles.
Please see the Research Environment User Guide for detailed documentation on how to use and query the Genomics England dataset. This page also includes instructional videos which can not be viewed from within the Research Environment.
Release overview¶
The NHS Genomic Medicine Service (GMS) Data Release Version 3 provides clinical data for 25,694 participants which are part of 14,982 referrals. In summary, this release includes 27,235 genomes from 25,300 participants. There are 23,667 genomes from 23,546 rare disease programme participants and 3,568 genomes from 1,760 cancer programme participants.
We further provide tiering data from 12,435 referrals, and 12,232 interpretations are represented in the GMC Exit Questionnaire of which 24.6% have a case_solved status of "yes". Within the GMC Exit Questionnaire, cases are included up to 20-02-2024. This release includes 13,026 interpretation requests from the Rare Disease program and 1,784 interpretation requests from the Cancer program.
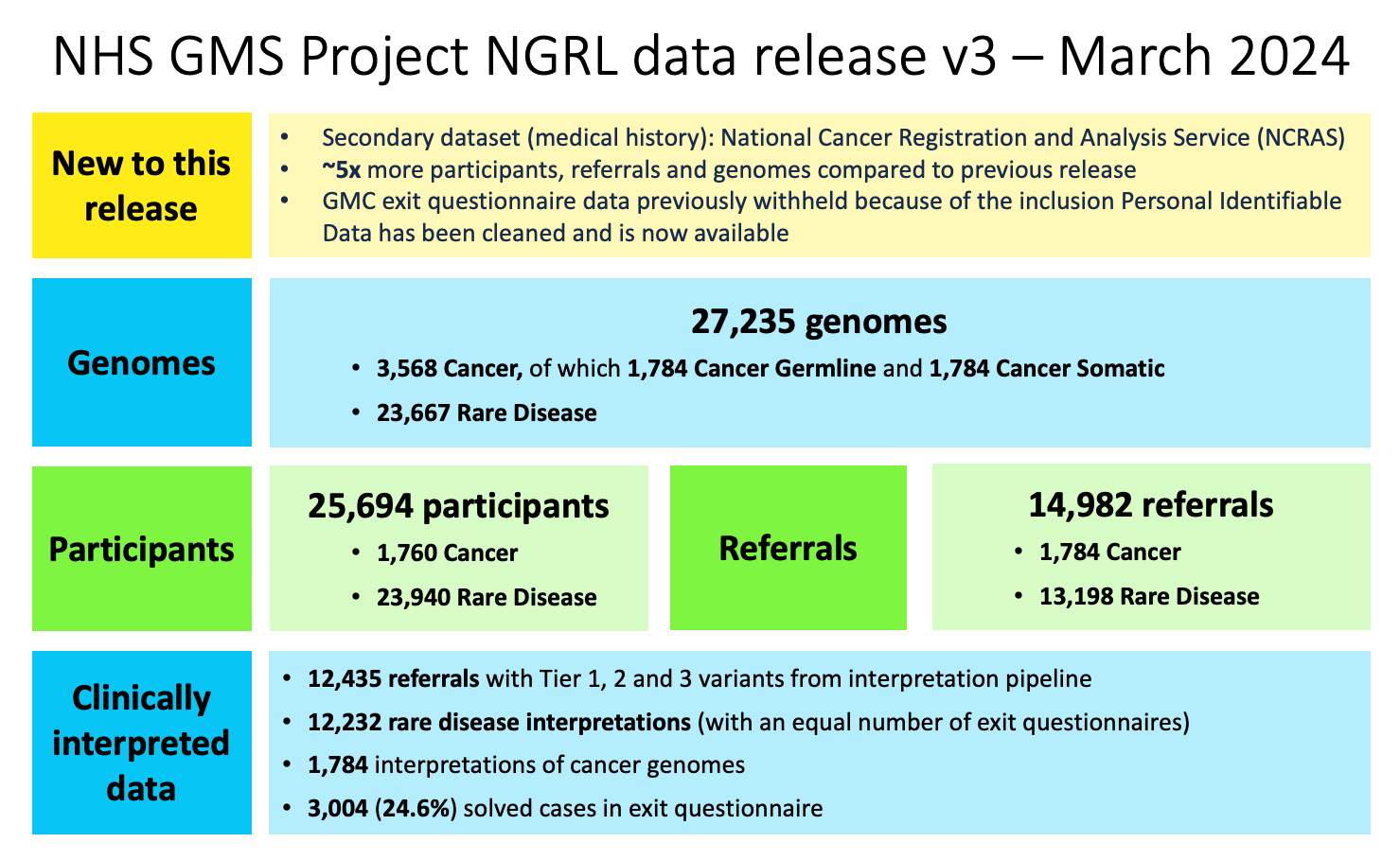
Table overview of genomic data:
| Type | Genomes count | Participant count |
|---|---|---|
| Rare Disease | 23,667 | 23,546 |
| Cancer Germline | 1,784 | 1,760 |
| Cancer Tumour | 1,784 | 1,760 |
| Cancer Total | 3,568 | 1,760 |
| Genomes Total | 27,235 | 25,300(*) |
Participants by program(*) breakdown:
| Programme | Participants | Referrals |
|---|---|---|
| Rare Disease | 23,940 | 13,198 |
| Cancer | 1,760 | 1,784 |
(*) Participants can be part of multiple referrals and across programs.
Audience¶
The intended audience for this document is researchers that have access to the Genomics England Research Environment.
Identifying this data release¶
The clinical data and tabulated bioinformatic data for this data release, and the paths to the applicable genome files, are found in the following LabKey folder:
nhs-gms-release_v3_2024-03-18
Subsequent releases will be identified by an incremental increase in the version number and the date of data release.
Relevant genomic data produced by the Genomics England Bioinformatics pipeline (i.e. joint-called VCFs, annotated somatic VCFs) can be found in the your home directory, under the folder gel_data_resources and then gms.
Scope¶
For release v3, the inclusion criteria are as follows:
- Participant has been through a manual consent audit and passed
- Only those participants who had all of their consent documents audited, and all documents consistently confirmed that they were eligible (they had both discussed and consented to inclusion in the NGRL, and were consented as an adult or child) are included.
- Any participants who were consented as children but were already 16 at the time of consent, or have since turned sixteen (but are not deceased) are deemed ineligible. Unless they have been reconsented as an adult.
- Participant is part of an eligible referral
- Eligible referrals refer to closed cases that contain at least one eligible participant.
In scope¶
Below we provide an overview of the data in scope for this release. By definition, this relates to cancer and rare disease data for participants enrolled in NHS GMS that consented for research. These data include:
- Genomic data for participants when available.
- This data contains closed case data only. This means that all referrals have gone through interpretation.
- Whole genome sequencing (WGS) family-based quality control for rare disease.
- Outputs of the Genomics England Bioinformatics rare diseases interpretation pipeline
- Tiering data – rare disease
- Exomiser results for interpreted genomes – rare disease
- GMC outcome data ("exit questionnaire data") – rare disease - up until 20/02/2024.
- Outputs of the Genomics England Bioinformatics cancer interpretation pipeline
- 'Gold standard' cancer genomes which have been through interpretation and passed quality checks
- Annotation and tiering of small variants
- Primary clinical data, including recruited disease and primary tumour types
- Secondary datasets (medical history) from National Cancer Registration and Analysis Service (NCRAS)
Out of scope¶
Additional time is required to update the applications/tools that are available in the RE to the current data release, e.g. IVA, Participant Explorer. Please refer to the Application Data Versions page for the data release version used in the RE products and services.
Data out of scope for this release:
- Clinical and genomic data for participants that have withdrawn from research after enrolment to the Genomic Medicine Service, or were otherwise ineligible.
- Participant data from the pilot phases of the 100,000 Genomes Project (i.e. not main programme).
- Participant data from the 100,000 Genomes Project (main programme).
Quality notes¶
This section will be amended for future releases as more documentation becomes available.
Note on Labkey platekey query limitations¶
Aggregation or Distinct queries including specifically the platekey column (e.g. SELECT DISTINCT participant_id, platekey FROM <table_name>) in Labkey will intentionally fail with a 'Status code = 500' error , UnauthorizedException or 'Unable to locate required logging column Key'. This can initially be circumvented by pulling in the entire data with SELECT * FROM <table_name> and subsetting or filtering your data downstream. We will continue to monitor the impact of the issue.
Terms of use for specific cohorts¶
For NHS GMS Data Release Version 3, no cohort has been formally linked to the data. This will change in future releases and the Terms of Use for Specific Cohorts will be amended.
Data release description¶
For an overview of the tables available in LabKey please see: NHS GMS dataset overview
The Genomics England data are organised into data views (displayed within LabKey as tables) categorised into common, bioinformatics and cancer. The data dictionary describes the table structure and provides data definitions for this release.
Change summary¶
The change summary for the NHS GMS data release can be found here: NHS GMS data release change summary
Contact and support¶
For all queries relating to this data release please contact the Genomics England Service Desk portal: Service Desk (accessible from outside the Research Environment). The Service Desk is supported by dedicated Genomics England staff for all relevant questions.