Extract variants for a list of genes¶
This workflow extracts variants of the specified gene region(s) from the 100kGP Illumina VCF files, aggregates them all together and annotates variants with VEP.
Description¶
A common query is to ask for the number of samples that have variants within a gene, or list of genes. This workflow supports that query.
The workflow is written in WDL and is run using the Cromwell engine. It relies on four external tools in order to execute the workflow. These are:
Please find more detailed information on various steps of the workflow in the "Further reading" section to learn more about them.
Briefly, the workflow, submitted to the final_output/data/".
Notes¶
- IMPORTANT: The VEP config file for annotations (see below, "Workflow Files") can be edited, but please note that PolyPhen2 scores cannot be used by our industry Research Network members unless their Company/Organisation separately purchase a commercial licence from PolyPhen2 - our Research Network members can instead freely use PolyPhen2 scores, simply add a tab-separated line in this file for that, similar to the SIFT line, that reads "polyphen b".
- The workflow will crash if the same the final output folder already contains files with the same file name. For example, if you ran the workflow for the BRCA1 gene on day one, and you do so unchanged on day two (for instance as part of a list of genes) it will crash trying to copy the final data to the output folder. This is done in order to protect your outputs from accidental overwriting - if you do not need a gene's file any more please remove it before re-running the workflow in the same location.
Please run the workflow on a small number of genes only. On average the workflow should complete within 2-3 hours per gene, however this largely depends on the gene size and on the traffic on our HPC. Therefore, sometimes the workflow may take up to a full day to complete, even on a small number of genes.
- In order to save disk space, please delete the content of output folders "
cromwell-workflow-logs" and "cromwell-executions" after you have checked the run was successful. Those folders simply contain redundant data and files that can be useful for debugging when the run fails. Additionally, the content of folders "final_output/call_logs" and "final_output/workflow_logs" can also be deleted safely after a successful run, if not needed.
Workflow files¶
The workflow files are located in:
/gel_data_resources/workflows/BRS_tools_geneVariantWorkflow/v1.7/
The workflow is contained within three files specified by the .wdl suffix. These are:
variant_workflow.wdl(main workflow script)annotation.wdl(a sub workflow script, scattering over genome build)quick_merge.wdl(a second sub workflow script, scattering over vcfs)
There is one file that specifies runtime inputs. This is:
variant_workflow_inputs.json(specifies runtime inputs such as the gene list location, and Release Programme Version)
There is another file that specifies remaining options for the workflow. This is:
variant_workflow_options.json(specifies output dir for overall output and output dir for log-files)
There is a file which specifies which genes need to be used (max ten genes). This is:
gene_list.txt
There is a file which specifies the VEP annotation that will be included (see IMPORTANT note above, in the "Description" section). This is:
vep_config_file.txt
There is a single shell script that is used to submit the workflow to the HPC. This is:
submit_workflow.sh(see instructions below)
There is a file detailing how Cromwell interacts with the LSF scheduler. This is:
cromwell.conf(this file should never be changed)
Finally, there is a file containing file-paths to retrieve the gene coordinates. This is:
GRCH38_GRCH37_files.txt(this file should never be changed)
Instructions¶
Overview¶
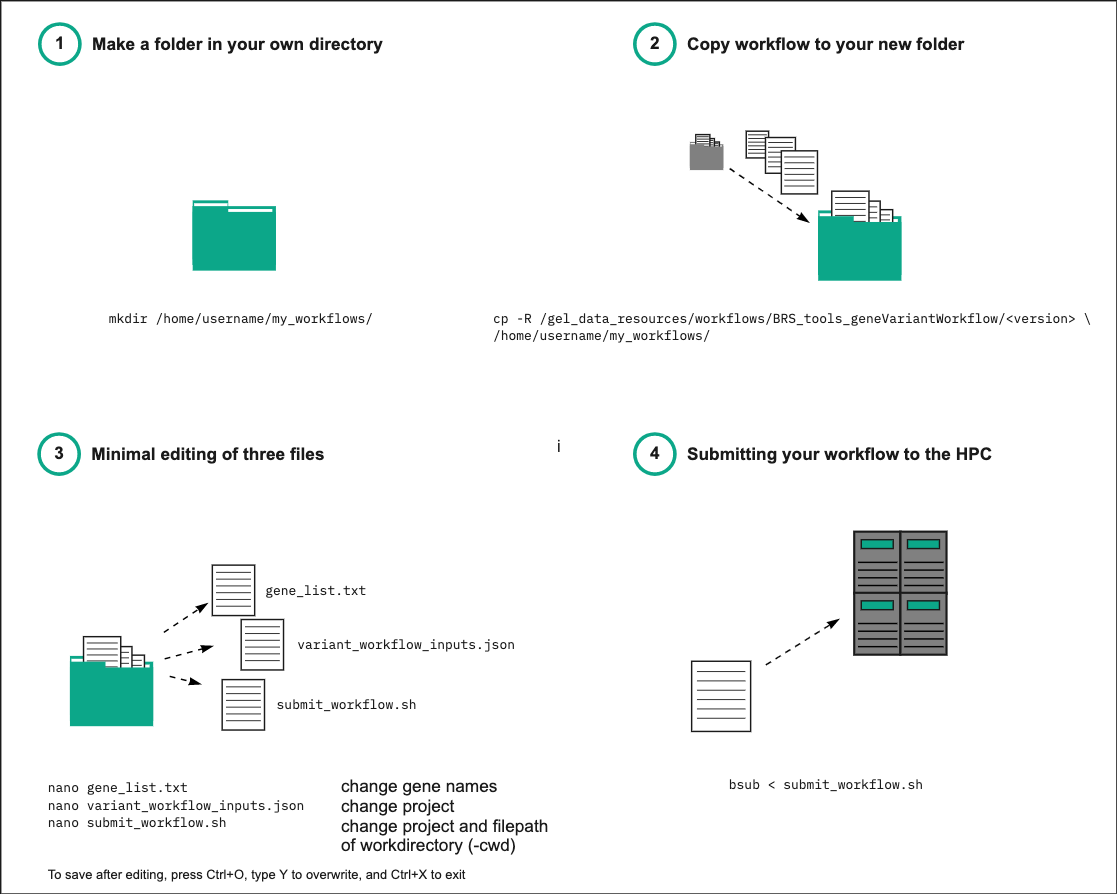
Above is a simplified schematic overview on what you need to do to run the script in four steps. This starts from creating a folder in your own work directory into which you can copy our files. It then points to a minimal number of files that you will need to edit and run the script. Below you will find a more detailed step-by-step description of each step involved (please note that the version in the file path mentioned in the figure above may not be accurate to the current situation).
Usage (tell me what I need to change to run it)¶
Before you start, please check whether you have configured your R LabKey API for the HPC. Most user feedback and troubleshooting linked issues to an incorrect setup of the R LabKey API. We have also changed the documentation based on this feedback to make it easier for all users to set it up without issue.
What to edit to run the workflow? And how to start it?
-
Copy all the files from our
/gel_data_resources/workflows/BRS_tools_geneVariantWorkflow/v1.7folder to your own workflow folder, andcdthere. -
Edit the
gene_list.txtinput file (a .txt file containing a list of HGNC gene symbols or Ensembl gene IDs) - please note that no more than 10 genes will be read and actioned. -
For example:
nano gene_list.txt -
Inside gene_list.txt add the gene names of interest (here BRCA1, TNF, and an Ensembl ID ENSG00000227518)
-
Save the file (Ctrl+O)
-
Edit both
submit_workflow.shandvariant_workflow_inputs.jsonto specify a valid project code for you (the default is set to "bio" in both files) and make sure that you are accessing the latest version of LabKey. -
OPTIONALLY, edit file
submit_workflow.shto specify the main working directory (the default is set to your current working directory) - note that in this case, all relative paths in config and input files need to refer to this directory: -
#BSUB -cwd /filepath/to/your/workflow_folder/ -
Please make sure you have read the IMPORTANT note in the "Description" section above.
-
Once you have edited all the above files accordingly, you are ready to run the script.
-
In the terminal (HPC), enter the following line:
bsub < ./submit_workflow.sh
Inputs and outputs¶
| Type | Type | File or Folder | Description | Requires editing |
|---|---|---|---|---|
| Input | File | gene_list.txt | .txt file containing a list of HGNC gene symbols or Ensembl gene IDs (new line separation) - no more than 10 genes will be read. | yes |
| Input | File | variant_workflow.wdl | Main workflow script. | |
| Input | File | annotation.wdl | A sub workflow script, scattering over genome build. | no |
| Input | File | quick_merge.wdl | A second sub workflow script, scattering over vcfs. | no |
| Input | File | variant_workflow_inputs.json | Specifies runtime inputs such as the gene list location, and Release Programme Version. | yes (for project code) |
| Input | File | variant_workflow_options.json | Specifies output dir for overall output and output dir for log-files. | no (optional) |
| Input | File | cromwell.conf | Configuration file on how Cromwell interacts with the LSF scheduler. | no |
| Input | File | GRCH38_GRCH37_files.txt | Contains the locations of files with gene identifiers and their respective regions. | no |
| Input | File | submit_workflow.sh | The final submission script to run the workflow. | yes (to latest database version, your project code and optionally working directory) |
| Input | File | vep_config_file.txt | VEP configuration file to specify how the data should be annotated. | no (optional) |
| Output | File | final_output/data/adjustedInput\({genome_version}_\).tsv_ | The file that is created based on your gene input. This is used further downstream in the workflow. Any duplicate regions are filtered out and placed in the entryDuplicate column (see below). | NA |
| Output | File | final_output/data/Genes_not_search\({genome_version}_\).tsv_ | The file that is created listing genes that could not be detected in the flat file containing all gene and Ensembl IDs (optional creation). | NA |
| Output | File | final_output/data/logfile${timestamp}.txt_ | File that is produced containing software and release programme version information. | NA |
| Output | File | final_output/data/\({chr_gene_ensemblID}_\)_annotated_variants.tsv | Main output file containing all the variants and their respective annotation. Each row will have corresponding platekey IDs attached. | NA |
| Output | File | final_output/data/\({chr_gene_ensemblID}_\)_result_left_norm_tagged_vcf.gz | VCF corresponding to the annotated_variants.tsv file. | NA |
| Output | File | final_output/data/\({chr_gene_ensemblID}_\)_result_left_norm_tagged_vcf.gz.tbi | VCF.tbi corresponding to the annotated_variants.tsv file. | NA |
| Output | Folder | final_output/call_logs/ | Contains all the log files that are generated during the query. | NA |
| Output | Folder | final_output/workflow_logs/ | Will contain the running workflow log. | NA |
| Output | Folder | cromwell-workflow-logs/ | Will contain the overall workflow log. | NA |
| Output | Folder | cromwell-executions/ | All the temporary files for each run are stored within the genetic_report folder. Once the jobs have been completed, they will be copied over to the final output folder specified by the user. This will also contain the cromwell database for callcaching. In case an identical query is being done, the workflow will check this database to see if it has been done before, and simply extract the data again without running the full script again. |
NA |
Output breakdown¶
Adjusted input file (adjustedInput\({genome_version}_\).tsv_):
| Column | Description |
|---|---|
| chr | chromosome |
| start | start position of gene region |
| end | end position of gene region |
| gene | HGNC gene symbol |
| ensemblID | Ensembl gene ID |
| internalID | identifier generated internally which will be used as a prefix for output files: chr_gene_ensemblID |
| posTag | position tag (chromosome_start_end) aiding the identifier deduplication step |
| originalInput | specifies what has been used as an input by the user, either "gene" or "ensembl", referring to either gene symbol or Ensembl ID |
| entryDuplicate | NA, or containing a comma-separated string of internalIDs. The internalIDs listed here are considered the same entry due to their identical genomic position. |
Genes not searched file (Genes_not_search\({genome_version}_\).tsv_):
Similar to the input file specifying the entries which were not detected in the flat file containing all gene and Ensembl IDs.
Variant output files \({chr_gene_ensemblID}_\)_annotated_variants.tsv):
| Column | Description |
|---|---|
| CHROM | Chromosome |
| POS | Variant position |
| ID_variant | Internal identifier of the variant for the workflow. This does not correspond to any identifiers in public databases |
| REF | Reference allele |
| ALT | Alternate allele |
| AN_variant | Total number of alleles in called genotypes |
| AC_variant | Allele count in genotypes |
| AC_Hom_variant | Allele counts in homozygous genotypes |
| AC_Het_variant | Allele counts in heterozygous genotypes |
| AC_Hemi_variant | Allele counts in hemizygous genotypes |
| MAF_variant | Allele frequency of the variant |
| NS_variant | Number of samples with data |
| DUP_variant | Number of duplicate variants |
| Various annotation columns (Impact and consequence annotation, variant class, feature type, ClinVar, gnomAD allele frequencies if existent, etc.) | |
| <_samples columns> | These contain comma separated platekey IDs for each of the variants referring to the Hom, Het, or Hemi genotypes. |
VCF output files
_ _result_left_norm_tagged_vcf.gz _ _result_left_norm_tagged_vcf.gz.tbi
Further reading¶
How it works¶
The first steps will fetch the coordinates of the gene inputs (HGNC gene symbols or Ensembl gene IDs) and the cohort with corresponding filepaths (fetch_coord and fetch_samples). The workflow will then run the variant collection in chunks of 1000 samples (split). Then, per chunk, retrieve all the associated vcf files and extract the variants according to the provided coordinates. These are then merged to a single bcf file. This is the first-round merge (first_round_merge). In the second-round merge, the output from the first-round are merged together. In this step, normalisation takes place to remove any duplicates, the index is created, and the internal variant identifier is created (second_round_merge). In the next phase, allele counts and MAF are calculated and plate-keys for participants associated to the type of variant are identified (fill_tags_query). Afterwards, the variant files can be annotated using VEP. The config file will specify which annotations are included (annotate). Finally, after annotation, the platekeys are added to the annotation file, and the annotation is filtered based on the input specifically for HGNC gene symbol or Ensembl gene ID (sum_and_annotate). Once these steps have been completed, the data is copied to the output directory specified by the user, or the default.
Adding custom annotation¶
By default, the workflow will annotate the relevant variants using Clinvar on top of the VEP annotations - see the vep_extra_configs variable inside file variant_workflow_inputs.json .
Further annotation can be added using that input variable, and specifying the annotation sources separately for GRCh37 and for GRCh38. For example, your "high_confidence" custom annotation files could be added by modifying the section for the vep_extra_configs variable as follows:
Also, the ClinVar version can be modified using the same input variable.
Task: fetch_coord¶
The gene or Ensembl IDs that are provided first undergo a screening process. The workflow uses a flat file for this screening process which is generated through BioMart for GRCh37 and GRCh38 containing the gene and Ensembl IDs, and gene regions (located in /public_data_resources/). The input containing your genes of interest will then be matched with the IDs present within this flat file, whether these are gene symbols, Ensembl IDs or a mix of both.
After the screening and the generation of an internalID (= chr_gene_ensemblID), a file is generated containing the gene regions, specific IDs, the internalID, type of input, and whether duplicate entries were detected (see below). This file is part of the final output, so you will always see what the actual workflow input was.
If the input cannot be detected, a separate output file is generated at the end containing the IDs for which no matching ID could be detected.
Handling duplicate IDs¶
It is possible that a single gene symbol will have multiple entries in the flat file, which have different Ensembl IDs. At this point, two things can occur:
-
if the regions are not identical, it will be considered as different entries, and therefore the workflow will consider them as separate entries, each output file receiving the unique internalIDs as a prefix. Or
-
if the region is identical, the other entries with the same gene symbol are deprecated, and the first entry is retained. The internalIDs of the deprecated entries are stored and placed in the “entryDuplicate” column of the primary entry. If different gene symbols have the same Ensembl ID, a similar process will take place.