Getting help¶
Encountering blockers within the Research Environment (RE) can be frustrating. To help you resolve some common issues, we've made some resources available, highlighted here. We recommend you take a look at these before you contact us, as this will be the quickest way to resolve your issue.
Getting started¶
For general enquiries, you may be interested in our pages on
What we can and can't help with¶
We can help you with using the RE tools, especially where there are configuration options specific to the RE. However we cannot provide any advanced training in:
- programming/scripting
- bioinformatics
- data science
- biology
Workflows¶
We have created a number of workflows for analysing our data, designed to be applicable for a wide range of research questions. Each workflow has extensive documentation which includes example data. We suggest a test-run with the example data before you get started on your own research.
There are also some recommendations for resolving commonly occurring problems. These recommendations are the steps we would use to fix or diagnose your issue, so you may find that you can fix it yourself this way, or at least provide us with more information if you do need to contact us. If you do contact us, please provide the outputs of any diagnosis steps.
Internal troubleshooting steps¶
We would usually consider the following questions:
- Is it a known issue with a workaround?
- Can we replicate the problem? We would test this with the example data first, then with your research data.
- Could this be a bug in the tool? If so does this bug affect multiple versions?
- If the error relates to file path access issue, we would check the source of the file path. It's possible that you're trying to access paths that are restricted.
- Are there other tools and software you need to load before you can run your job?
- Is the analysis trying to access a resource that is not whitelisted in the Research Environment?
- Is there a section of the User Guide that covers your use-case? Check to see if what you're trying to do is actually possible.
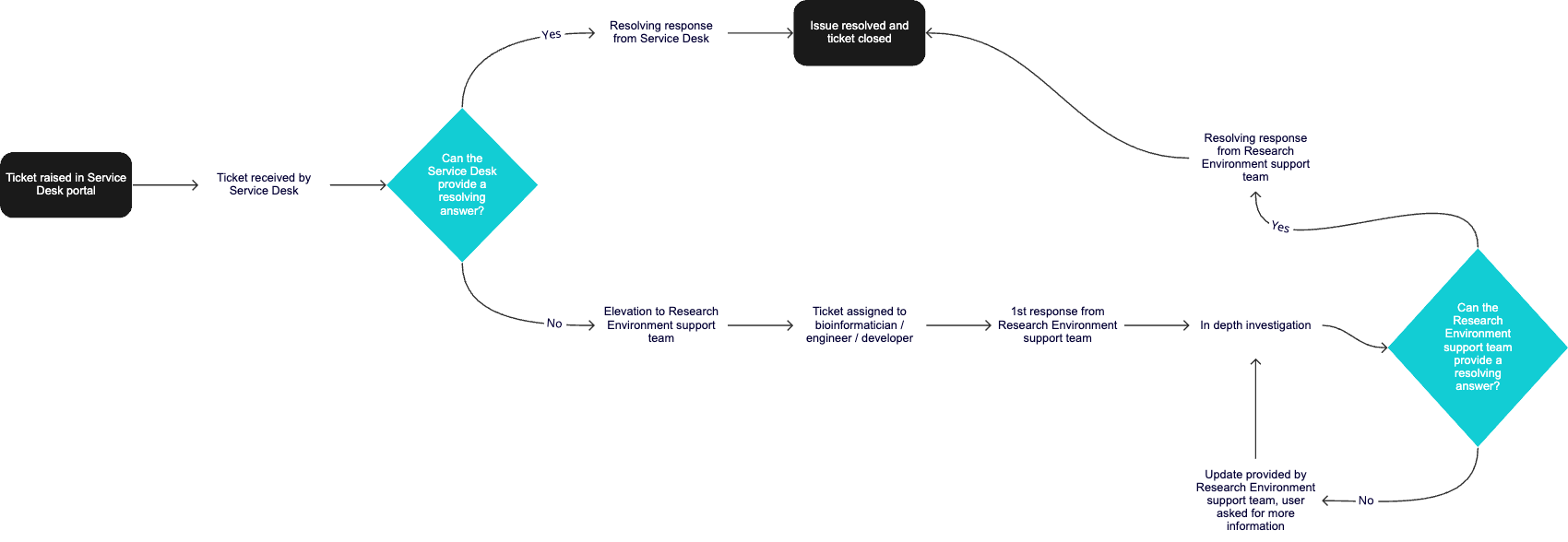
If you have considered these points and still not managed to resolve your problem, please raise a support ticket including information about all the troubleshooting you've done.
Contacting the Service Desk¶
If you still need help, then please log an issue in the Genomics England Service Desk Portal. If this is your first time using the Service Desk, you will need to create an account first as your Research Environment credentials will not work. You can monitor your tickets via the 'Requests' link on the portal.
General information to include¶
- Research Network or commercial
- your username
- which environment you are using (Research Environment, Cloud-RE, HPC)
- a brief description of the problem
- relevant error codes
- tests you've done and their results
- how to reproduce the error
- recent changes you've made to your configuration
- a screenshot of the error (avoid including participant data)
Usually, you are not allowed to take screenshots within the RE, however you can send these to the Genomics England Service Desk for the purpose of help requests, where relevant. In this case, please redact any identifiable data and purge the screenshots from your system as soon as you create the ticket. Where it's necessary to include participant level data, we will provide you with a location in the RE to share the data.
Help us to help you¶
To prevent a lot of early back-and-forth, here are some of the things we need to know to help resolve your issue. Please include them in your ticket. We've split this section into different types of issues.
Login issues¶
If you are experiencing login issues in the Research Environment:
- If you are unable to log into the RE generally, please mention the RE specifically.
- If you cannot log into an application, please state which one you are attempting to access.
- Many login problems are resolved by "refreshing" the session, so it is good practice to ensure all your files are saved so no progress is lost.
Workflows, analysis tools and coding errors¶
For these ticket types, it is extremely useful to have access to your working directory:
- For Research Network users:
- Raise a Service Desk ticket to generate a GEL-XXXXX reference number
- Copy your working directory files to a subfolder named with your GEL reference number (as above) in /re_gecip/shared_all_gecips (ensuring the folder allows us read permissions
chmod 755 -R GEL_XXXXX/)
- For Commercial users:
- Raise a Service Desk ticket to generate a GEL-XXXXX reference number
- We do not yet have a location for you to privately share your files with us and not other commercial users. Please instead provide screenshots of your code and errors in the ticket.
Data queries¶
- For data access issues:
- Is this the first time you have tried to access this data? If not, has something changed?
- What is the source of the file_paths you are using?
- For requesting data to be made available in the TRE:
- How large is the data being requested (MB/GB)?
- Are there any license requirements?
- Can you share any links to help us assess the suitability of the data?
Software requests and whitelisting¶
GEL has a duty of care towards our participants who have generously shared their data. We will not whitelist any websites if there is a chance that this will allow data to exit the RE.
For software requests you should include:
- For all software, please state the license covering the software, and whether this license permits use by academic and/or commercial use. This must meet our licensing policy.
- For R packages, please let us know which version of R you're using – this will allow us to provide the package in the correct modules. Other relevant information will be requested within the Software Request ticket
- For python packages, we recommend creating a personal conda environment. However, a limited number of conda channels are whitelisted; if you need a package from a non-whitelisted channel, you will need to raise a Software Request ticket.
For whitelisting requests you should include:
- links to the resource itself, plus any documents that help with making the approval decision.
- why the resource will be beneficial.
- what data will be entering the RE and potentially leaving the RE via the proposed website.
HPC usage and job submission¶
- What node are you working on? You will be able to get this from the command prompt of your HPC session or the "Sender" line of your LSF standard out file (for example,
Sender: LSF System <lsfadmin@phpgridzlsfe020>) - Your submission script, according to the instructions outlined in Workflows, analysis tools and coding errors
Applications (e.g. Labkey, Participant Explorer...)¶
To ensure tickets of this nature reach the correct support teams, please provide as much of the following information as possible:
- the user guide pages you are following.
- the expected behaviour compared to the one you receive/observe.
- Are you requesting help or reporting an error?
- If the issue relates to logging in, please check whether you are able to log into other applications.
- Is this the first time you have tried to complete this task? If not, has something changed?
- links and scripts for any API usage.
How we triage your tickets¶