IVA Catalog¶
The IVA Catalog allows you to browse the metadata stored in OpenCGA. In this case, metadata is all the information that doesn't correspond to genotypic information or the variants per se. You can use Catalog to explore the individuals, samples and families whose variants are stored in the database by searching with IDs, phenotypes, disorders, processing etc.
For more information about the data models for each entity, please refer to the official Data Models OpenCGA documentation.
Accessing catalog¶
You can access by clicking Catalog in the upper bar in IVA. A drop down will be shown with the different browsers as showed below:

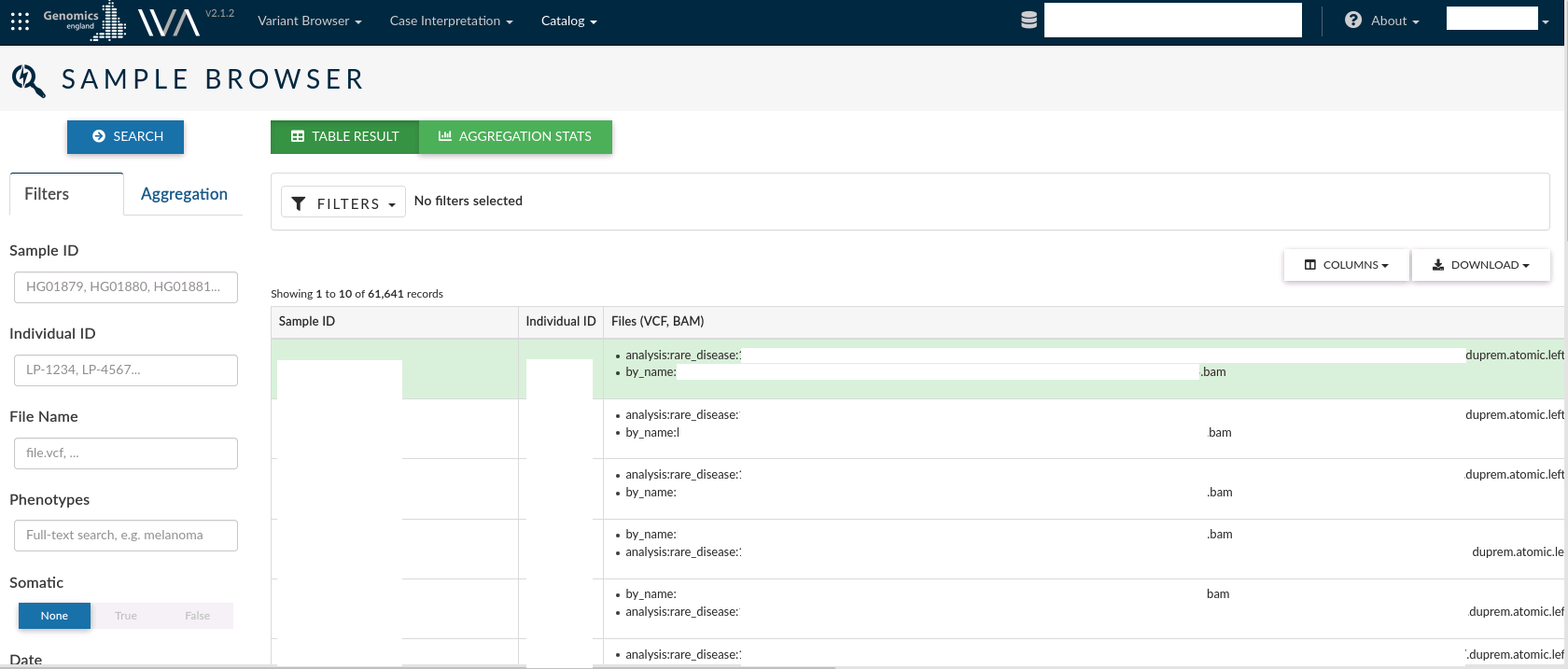
Sample browser¶
The sample browser allows you to search over all the records information for each sample in a study. For each Sample, you can see the Sample id, the individual ID from which the sample was taken and the output files from the analysis conducted for the sample. The VCFs associated with each sample are the files from which the variants are extracted to load into OpenCGA.
The left menu allows you to filter samples by Sample ID, Individual ID, File Name or Phenotypes. You can also select the somatic or germinal samples, or filter by the date the samples were created.
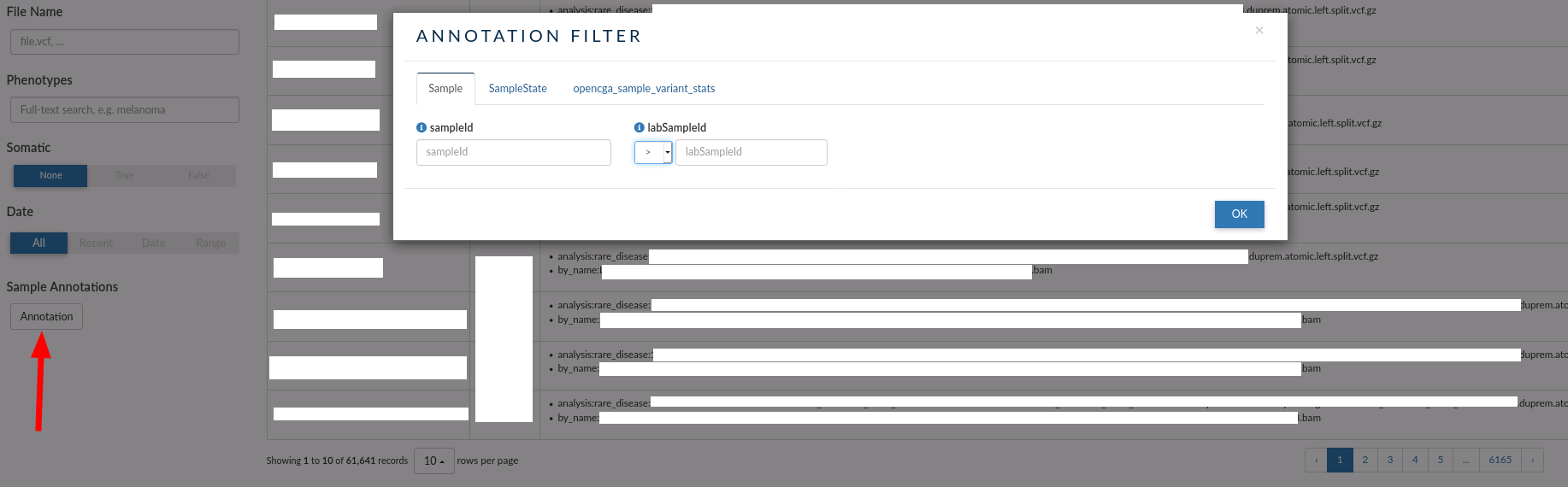
Sample annotations¶
IVA also allows you to filter by custom annotations. Those are attributes that are not defined in the sample data model but that can be specified by the owner of the study.
In this case, there are three custom annotations that you can use for browsing over the samples:
- Samples: You can filter by sample Id or lab sample Id (the ID used by the laboratory when the sample was processed).
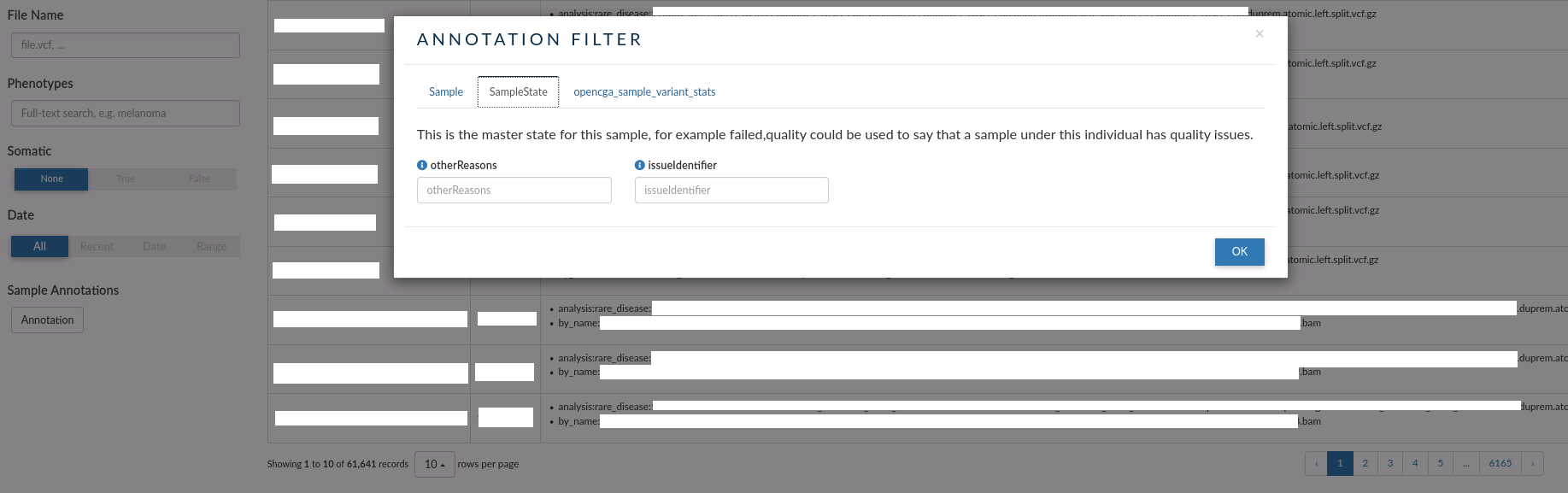
- SampleState: this tab allows to filter by issue with the samples. Eg: failed, quality etc.
- opencga_sample_variant_stats: you can use this tab to filter samples by their global statistic values over the study where the samples are found.
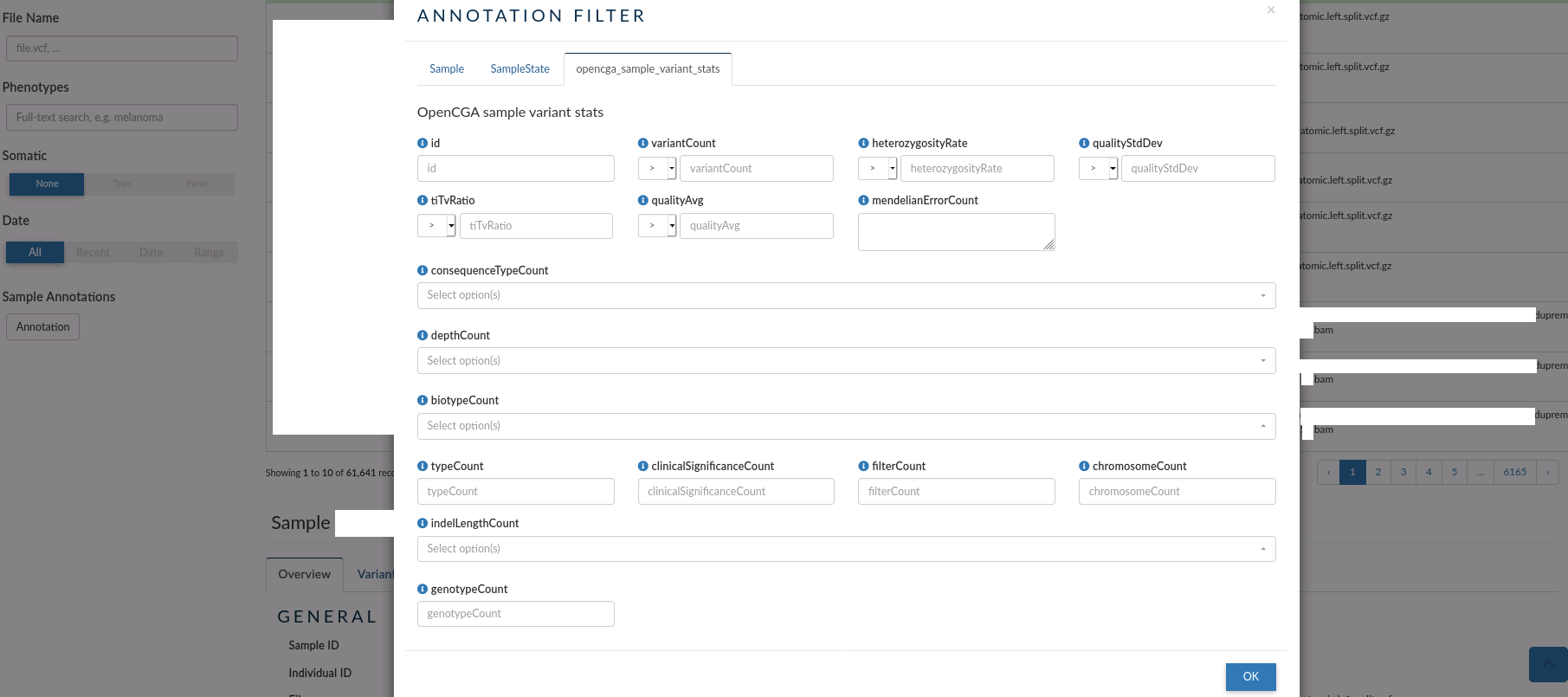
Individual browser¶
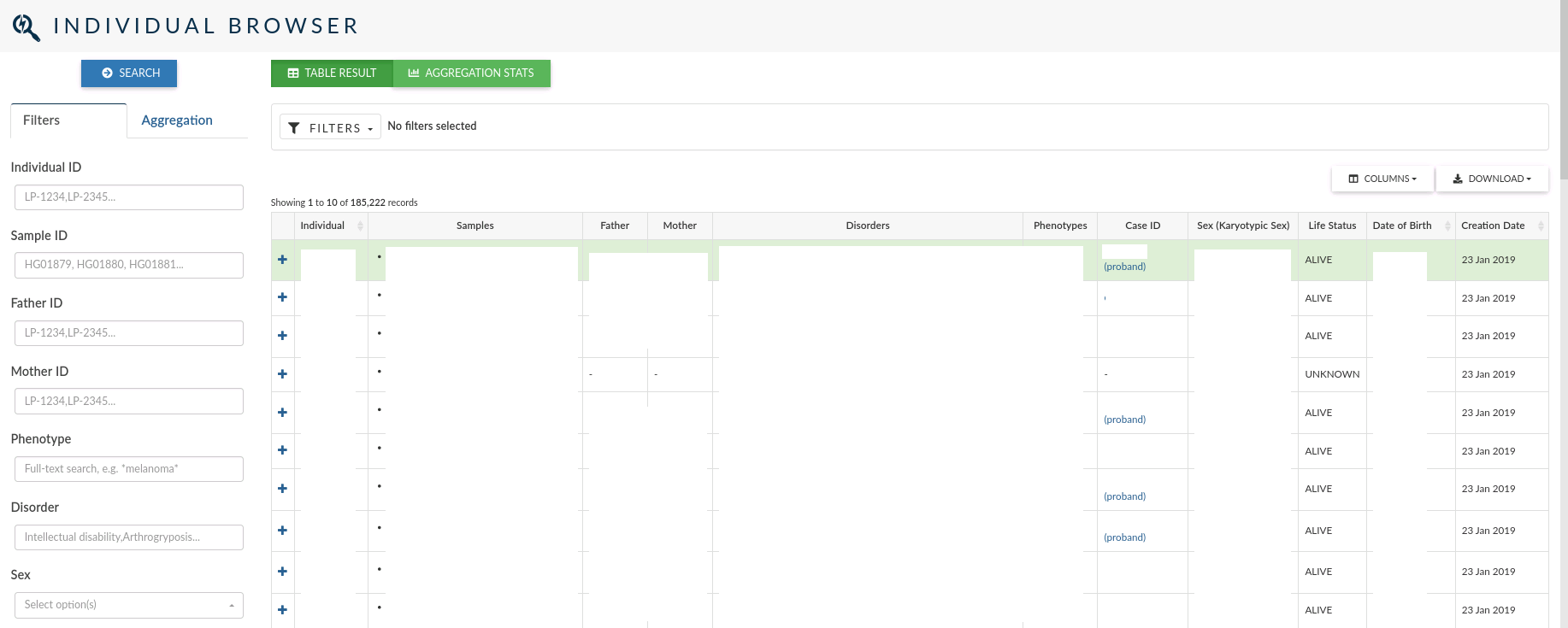
The individual browser allows you to search over all the records for the individuals which samples have been analysed in a study.
In the main table, for each Individual, you can see the id, the samples associated to the individual, the disorders and phenotypes assigned, the clinical case open for the individual (if it exists) the sex, life status, date of birth and creation date.
The left menu allows you to filter individuals by: Individual ID, Sample ID, Father and Mother IDs, Phenotype, Disorder, Sex, Karyotypic Sex, Ethnicity and Life Status . You can also filter by the date the individuals were created.
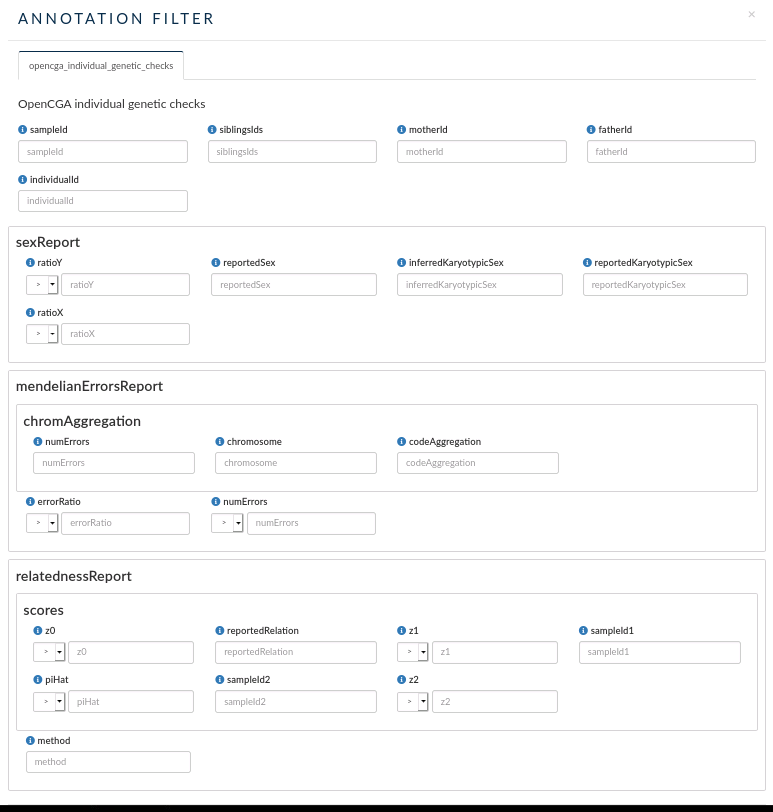
Individual annotations¶
Same as with samples, IVA also allows you to filter by individual custom annotations. In this case, there is just one annotation:
- opencga_individual_genetic_checks: this custom annotation allows you to search individuals by their mother, father or sibling Ids.
- You can also filter by other parameters defined by some individual stats:
- sexReport: filter by ratio of Y and/or X chromosome, reported and inferred Karyotypic Sex and reported Sex.
- Mendelian errors report: you can filter by errorRatio or number of errors. You can also filter by some parameter obtained from the chromosome aggregation.
- relatednessReport: you can filter individuals by some parameter obtained by performing a relatedness analysis between the individuals in the OpenCGA Catalog database.
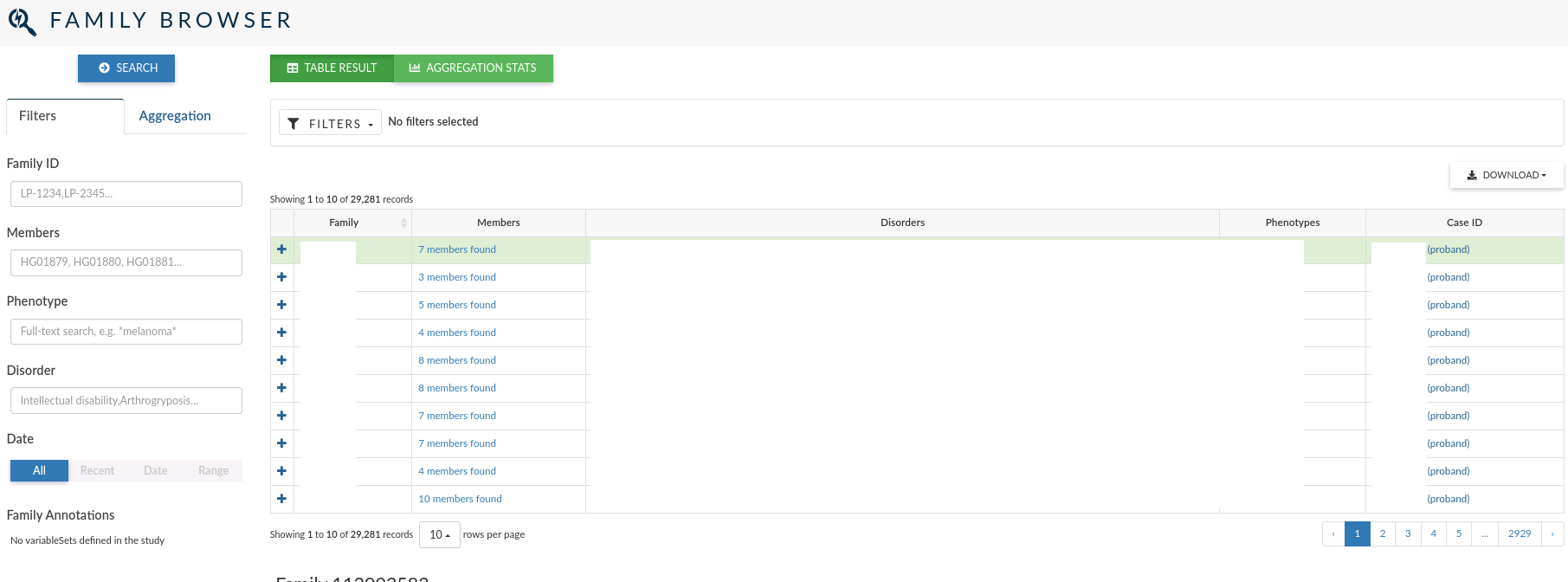
Family browser¶
The family browser allows you to search over all the records for the family that have been defined to depict the relations between the individuals whose genomes have been analysed.
In the main table, for each family, you can see the family id, the members who compose the family, the disorders and phenotypes assigned and the clinical case ID open for the family (if it exists).
The left menu allows you to filter individuals by: Family ID, Members, Phenotype and Disorder. You can also filter by the date the family was created.
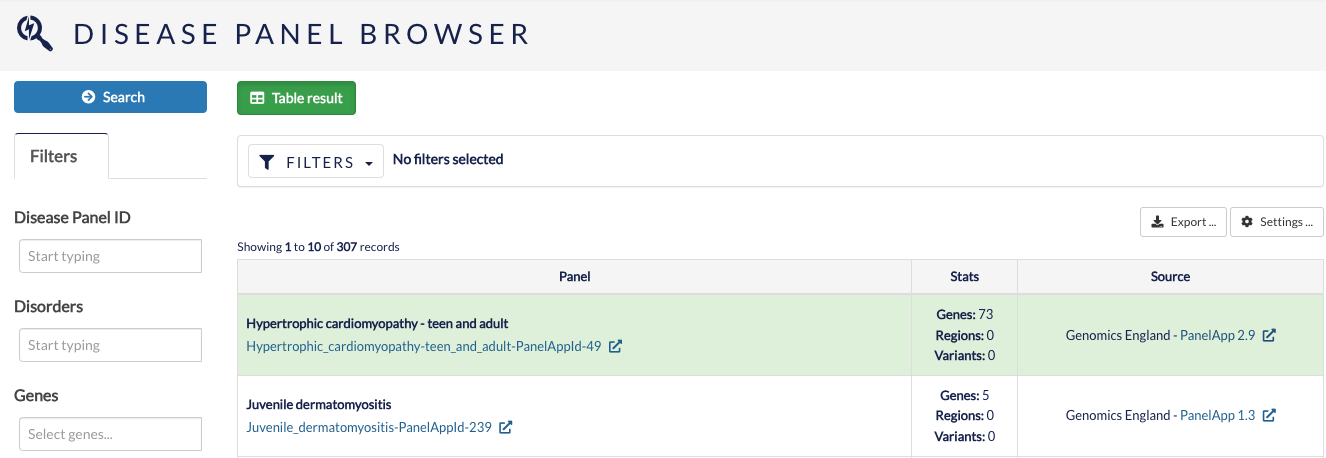
Disease panel browser¶
The disease panel browser allows you to search for gene panels by IDs, disorders, genes and loci.