Data in Participant Explorer¶
Data sources¶
Participant data¶
Participant clinical data is obtained from the source data in the most recent versions of the 100kGP data release and NHS_GMS data release. Please see the Participant Explorer release notes for the current data versions.
The source data are imported into a Postgres database and mapped to a standard data model (HL7 FHIR) using SQL. The Participant Explorer UI operates on top of the FHIR model, but hides the technical details of FHIR (such as element names and extension URLs) to create a user-friendly, intuitive interface. A detailed overview of the mapping from source tables and columns to elements in the UI is given below.
Various elements in the Participant Explorer UI also provide clickable deep links to the source data into LabKey.
Reference data¶
Terminology reference data is provided by a FHIR terminology server, developed by the AEHRC. For details of the terminology server instance we are using, see the Terminology Server page.
Conceptual data model¶
The diagram below depicts the clinical data model used for representing participant data in the Participant Explorer. This model is based on the HL7 FHIR resource model.
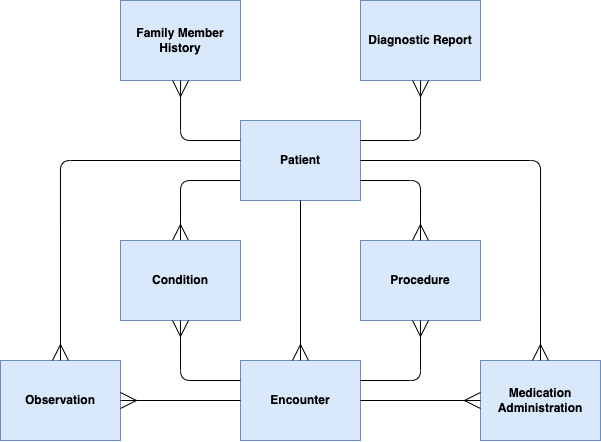
| Participant Explorer Label | FHIR Resource Type | Definition (from FHIR) | Meaning in terms of the 100kGP/NHS-GMS datasets |
|---|---|---|---|
| Participant | Patient | Demographics and other administrative information about an individual receiving care or other health-related services. | All consenting participants including probands and their relatives. |
| Referral | ServiceRequest | A record of a request for service such as diagnostic investigations, treatments, or operations to be performed | The referral to the Genomics Medicine Service for a genomic test for a specific clinical indication (NHS-GMS) or the recruitment to the 100kGP project for one of the eligible diseases. |
| Condition | Condition | A clinical condition, problem, diagnosis, or other event, situation, issue, or clinical concept that has risen to a level of concern. | Diagnoses from primary clinical data (including the recruited disease) and secondary data. |
| Observation | Observation | Measurements and simple assertions made about a patient | Phenotypic observations (HPO terms), Tumour morphology and stage observations |
| Procedure | Procedure | An action that is or was performed on or for a patient. This can be a physical intervention like an operation, or less invasive like long term services, counselling, or hypnotherapy. | Procedure and operation codes from primary and secondary data |
| Encounter | Encounter | An interaction between a patient and healthcare provider(s) for the purpose of providing healthcare service(s) or assessing the health status of a patient. | Grouping of conditions, observations and procedures by visit/event |
| Family Member | FamilyMemberHistory | Significant health conditions for a person related to the patient relevant in the context of care for the patient. | Key details and affected-status of family members of rare disease probands (Note: family members who are also participants will also have a patient/participant record) |
| Genome Sequence | DiagnosticReport | The findings and interpretation of diagnostic tests performed on patients, groups of patients, devices, and locations, and/or specimens derived from these. | Sequencing report meta information |
| Family Case Report | DiagnosticReport | The findings and interpretation of diagnostic tests performed on patients, groups of patients, devices, and locations, and/or specimens derived from these. | GMC exit questionnaire (case status and additional comments) |
| Drugs, Drug Group | MedicationAdministration | Describes the event of a patient consuming or otherwise being administered a medication. | SACT (chemotherapy) drug administrations |
Code systems overview - using the right codes¶
The following table can help you select the code systems to use when searching by clinical concept, depending on your area of interest and scope. It also indicates whether the "include mapped concepts" feature may be of use. Below the table are examples involving each code system.
Note that OMIM and ORPHA codes, present in NHS-GMS referral data, are not yet searchable in Participant Explorer so are not described here. However, they are displayed in the Participant Details and Download pages.
| Code System Short Name | Description | Primary Clinical Data (data associated with referrals) | Secondary Data (longitudinal data independent of referrals) | Concept Maps Available? |
|---|---|---|---|---|
| 100kGP Rare Disease | Rare disease groups, subgroups and specific diseases for which participants were recruited in the Genomics England 100kGP | Rare diseases groups, subgroups and specific diseases (100kGP only) | N/A | No |
| 100kGP Cancer Type | Cancer disease types for which participants were recruited in the Genomics England 100kGP | Cancer disease types (100kGP only) | N/A | No |
| 100kGP Cancer Subtype | Cancer disease subtypes for which participants were recruited in the Genomics England 100kGP | Cancer disease subtypes (100kGP only) | N/A | No |
| NHS-GMS Clinical Indication | Clinical indications for which participants were referred to the NHS-GMS | Referral clinical indications (NHS-GMS only) | N/A | No |
| ICD10 | ICD-10, WHO International Classification of Diseases | Cancer diagnoses (100kGP only) | Yes, SNOMED to ICD10 | |
| HPO | Human Phenotype Ontology | Observed phenotypes (rare disease referrals, "present" phenotypes only) | N/A | Yes, SNOMED to ICD10 |
| ICDO | ICD-O-3, WHO International Classification of Diseases for Oncology | Tumour morphology and topography (100kGP only) | NCRAS tumour morphology | No |
| OPCS | OPCS-4 Classification of Interventions and Procedures | Cancer imaging and surgery (100kGP only) | Yes, SNOMED to OPCS | |
| SNOMED | SNOMED CT (UK Edition) | Yes, SNOMED to ICD10, OPCS and HPO. | ||
| SACT Drug Group | Drugs used in treatments recorded in the SACT data | N/A | NCRAS chemotherapy drugs | No |
Examples¶
100kGP Rare Disease | Intellectual Disability: selects participants who were recruited in the 100kGP for intellectual disability (including affected relatives)
100kGP Cancer Type | Lung: selects participants who were recruited in the 100kGP for lung cancer.
NHS-GMS Clinical Indication | R29: Intellectual Disability: selects participants who were referred to the NHS-GMS with a clinical indication of intellectual disability (including affected relatives)
HPO | HP:0012622: Chronic kidney disease: selects participants with an observed phenotype of chronic kidney disease in their 100kGP or NHS-GMS referral (rare disease referrals only, including relatives, "present" phenotypes only).
ICD10 | C50: Malignant neoplasm of breast: selects participants with a diagnosis of breast cancer, either in their referral data or longitudinal data (this can include participants who are referred for a rare disease or other cancer types).
OPCS | J01: Transplantation of liver: selects participants with a liver transplantation record in their referral data or longitudinal data.
SCT | 241620005: Cardiac MRI: selects participants with a record of a cardiac MRI in their referral data or longitudinal data.
SCT | 38341003: Hypertensive disorder (with mapped concepts DISABLED): select participants with a diagnosis of hypertensive disorder in their longitudinal emergency care or mental health services data, because SNOMED diagnoses codes are currently only available in these data sets.
SCT | 38341003: Hypertensive disorder (with mapped concepts ENABLED): adds equivalent ICD10 and HPO codes for hypertensive disorder to the search criteria, and consequently selects participants with a record of hypertension in their referral data or longitudinal data. Disclaimer: the concept maps underlying this feature are not complete and can be inaccurate. Please review the included mapped concepts carefully, when using this feature.
ICDO | 80109: Carcinomatosis plus SCT | 307593001: Carcinomatosis: selects participants with this tumour morphology in their referral data or longitudinal data (this can include participants who are referred for a rare disease or other cancer types). Because tumour morphology/topography may be coded with ICD-O or SNOMED, it is advised to include both code systems when searching for a specific tumour morphology or topography.
Mapping of 100kGP and NHS-GMS data releases to data in the Participant Explorer¶
The tables below show how each data element in Participant Explorer is obtained from source tables in LabKey, including a description of any normalisation or filters applied in the Notes column.
Note: when downloading condition, observation, procedure or drug codes from Participant Explorer, the "Source Code" column contains the original source value, pre-normalisation.
For detailed information on tables and columns in the 100kGP and NHS-GMS data releases, please refer to the relevant data dictionaries of the 100kGP Data Release / NHS_GMS data release, and the Participant Explorer release notes for the data release versions available in Participant Explorer.
Participant¶
| Participant Explorer | Source Dataset | Source Table | Source Column | Notes |
|---|---|---|---|---|
| Participant ID | Both | participant | participant_id | |
| Year of Birth | 100kGP | participant | year_of_birth | |
| NHS-GMS | participant | participant_year_of_birth | ||
| Stated Gender | 100kGP | participant | participant_stated_gender | |
| NHS-GMS | participant | administrative_gender | ||
| Phenotypic Sex | 100kGP | participant | participant_phenotypic_sex | |
| NHS-GMS | participant | phenotypic_sex | ||
| Ethnic Category | 100kGP | participant | participant_ethnic_category | |
| NHS-GMS | participant | ethnicity_description | ||
| Life Status | 100kGP | death_details | death_date | if different from mortality, the value from mortality is used |
| NHS-GMS | participant | participant_year_of_death | if different from mortality, the value from mortality is used | |
| Both | mortality | date_of_death | takes precendence |
Referral¶
| Participant Explorer | Source Dataset | Source Table | Source Column | Notes |
|---|---|---|---|---|
| Referral ID / Family ID | 100kGP | participant | rare_diseases_family_id / gel_case_reference | Rare Disease / Cancer |
| NHS-GMS | referral | referral_id | Both Rare Disease and Cancer | |
| Proband/Relative | 100kGP | participant | participant_type | |
| NHS-GMS | referral_participant | referral_participant_is_proband | ||
| Disease Category/ Programme | 100kGP | participant | programme | |
| NHS-GMS | referral | category | ||
| Clinical Indication/ Recruited Disease | 100kGP | rare_diseases_participant_disease | normalised_specific_disease | |
| 100kGP | cancer_participant_disease | cancer_disease_type + cancer_disease_sub_type | ||
| NHS-GMS | referral | clinical_indication_full_name | ||
| Family Case Solved | 100kGP | gmc_exit_questionnaire | case_solved_family | |
| NHS-GMS | report_outcome_questionnaire | case_solved_family | ||
| Referral Date | 100kGP | N/A | Not populated for 100k, as for families the choice of date is unclear. In practice other registration events on the timeline show the likely period for the referral. | |
| NHS-GMS | referral | date_submitted | ||
| Family Members Tested | 100kGP | rare_diseases_family | family_group_type | |
| NHS-GMS | referral_test | referral_test_expected_number_of_participants | ||
| Family Members Available | 100kGP | participant | N/A | count of participants with family id |
| NHS-GMS | referral | N/A | count of participants with referral id |
Referral Members/Family Details¶
| Participant Explorer | Source Dataset | Source Table | Source Column | Notes |
|---|---|---|---|---|
| Relationship to Proband | 100kGP | rare_diseases_pedigree_member | father_id, mother_id | First-degree relationships are derived from father_id and mother_id. Others are displayed as "Family Member". |
| NHS-GMS | referral_participant | relationship_to_proband | ||
| Affection Status | 100kGP | rare_diseases_pedigree_member | affection_status | |
| NHS-GMS | referral_participant | disease_status | ||
| Stated Gender | 100kGP | participant | participant_stated_gender | not available for non-participants |
| NHS-GMS | participant | administrative_gender | ||
| Phenotypic Sex | 100kGP | participant / rare_diseases_pedigree_member | phenotypic_sex, father_id, mother_id | rare_diseases_pedigree_member entry is used for non-participants, otherwise participant table participant_phenotypic_sex overrides. |
| NHS-GMS | participant | phenotypic_sex | ||
| Participant ID | 100kGP | rare_diseases_pedigree_member | participant_id | |
| NHS-GMS | referral_participant | participant_id | ||
| Pedigree Member ID | 100kGP | rare_diseases_pedigree_member | rare_diseases_pedigree_member_id | only available for 100kGP |
Rare disease family case report¶
| Participant Explorer | Source Dataset | Source Table | Source Column | Notes |
|---|---|---|---|---|
| Family Case Comments | 100kGP | gmc_exit_questionnaire | additional_comments | |
| NHS-GMS | report_outcome_questionnaire | additional_comments | ||
| Position of "Family case report" on the timeline | Both | gmc_exit_questionnaire or report_outcome_questionnaire | event_date | |
| Medical Review Date | 100kGP | rare_diseases_family | family_medical_review_date |
Genome sequence report¶
| Participant Explorer | Source Dataset | Source Table | Source Column | Notes |
|---|---|---|---|---|
| Delivery ID | Both | sequencing_report | delivery_id | |
| Plate Key | Both | plate_key | ||
| Type | Both | type | ||
| Delivery Version | Both | delivery_version | ||
| Genome Build | Both | genome_build | ||
| Delivery Date | Both | delivery_date | ||
| Path | Both | path | ||
| Sample Date | 100kGP | clinic_sample | clinic_sample_datetime | sequencing_report is linked to clinic_sample via the lab_sample_id and clinic_sample_sk in the laboratory_sample table |
| NHS-GMS | sample | collection_date | sequencing_report is linked to sample via the referral_id |
Condition¶
| Participant Explorer | Source Dataset | Source Table | Source Column(s) | Code System | Notes |
|---|---|---|---|---|---|
| Condition Code | Both | av_tumour | site_icd10_o2 | ICD-10 | Normalised1 |
| 100kGP | cancer_invest_sample_pathology | primary_diagnosis_icd_code | ICD-10 | Normalised1 | |
| 100kGP | cancer_participant_disease | cancer_disease_type cancer_disease_sub_type |
Genomics England | ||
| 100kGP | cancer_participant_tumour | diagnosis_icd_code | ICD-10 | Normalised1 | |
| 100kGP | cancer_registry | cancer_site | ICD-10 | Normalised1 | |
| NHS-GMS | condition | code | OMIM or ORPHANET | ||
| Both | ecds | diagnosis_code_1 - diagnosis_code_12 | SNOMED CT | if diagnosis_qualifier_n = 410605003 ("confirmed") | |
| Both | hes_apc | diag_01 - diag_20 | ICD-10 | Normalised1 | |
| Both | hes_op | diag_01 - diag_12 | ICD-10 | Normalised1 | |
| 100kGP | mhmd_v4_event mhldds_event |
ic_eve_primarydiagnosis ic_eve_secondarydiagnosis |
ICD-10 ICD-10 |
Normalised1 | |
| 100kGP | mhsds_medical_history_previous_diagnosis | prevdiag diagschemeinuse |
ICD-10 / SNOMED | Normalised1 If diagschemeinuse is 1, 2, 02, ID, 4, 6 or 06 |
|
| 100kGP | mhsds_provisional_diagnosis | provdiag diagschemeinuse |
ICD-10 / SNOMED | Normalised1 If diagschemeinuse is 1, 2, 02, ID, 4, 6 or 06 |
|
| 100kGP | mhsds_primary_diagnosis | primdiag diagschemeinuse |
ICD-10 / SNOMED | Normalised1 If diagschemeinuse is 1, 2, 02, ID, 4, 6 or 06 |
|
| 100kGP | mhsds_secondary_diagnosis | secdiag diagschemeinuse |
ICD-10 / SNOMED | Normalised1 If diagschemeinuse is 1, 2, 02, ID, 4, 6 or 06 |
|
| 100kGP | mhsds_care_activity mhsds_indirect_activity |
codefind findschemeinuse |
ICD-10/ SNOMED | If diagschemeinuse is 1, 2, 02, ID, 4, 6 or 06 | |
| 100kGP | rare_diseases_participant_disease | normalised_disease_group normalised_disease_sub_group normalised_specific_disease |
Genomics England | ||
| Both | mortality | icd10_underlying_cause | ICD-10 | Normalised1 | |
| Both | mortality | icd10_multiple_cause_code_1 ... 15 | ICD-10 | Normalised1 | |
| Both | rtds | radiotherapydiagnosisicd | ICD-10 | Normalised1 | |
| Both | sact | primary_diagnosis | ICD-10 | Normalised1 | |
| Condition Source Code | Both | ||||
| Body Site Code | 100kGP | cancer_invest_sample_pathology | topography_snomed_ct_code | SNOMED CT | |
| TNM Stage Group | 100kGP | cancer_participant_tumour | integrated_tnm_stage_grouping | ||
| 100kGP | cancer_participant_tumour | ajcc_stage | |||
| Both | sact | sact_stage_at_start | |||
| Stage Best | Both | av_tumour | stage_best | ||
| Stage Best System | Both | av_tumour | stage_best_system | ||
| T Stage | Both | av_tumour | t_best | ||
| 100kGP | cancer_participant_tumour | component_tnm_t | |||
| M Stage | Both | av_tumour | m_best | ||
| 100kGP | cancer_participant_tumour | component_tnm_m | |||
| N Stage | Both | av_tumour | n_best | ||
| 100kGP | cancer_participant_tumour | component_tnm_n | |||
| Dukes | Both | av_tumour | dukes | ||
| 100kGP | cancer_participant_tumour | modified_dukes_stage | |||
| FIGO | Both | av_tumour | figo | ||
| 100kGP | cancer_participant_tumour | final_figo |
Observation¶
| Participant Explorer | Source Dataset | Source Table | Source Column(s) | Code System | Notes |
|---|---|---|---|---|---|
| Observation Code | Both | av_tumour | histology_coded | ICD-O-3 | |
| Both | av_tumour | stage_best | STAGE | ||
| Both | av_tumour | figo | STAGE | ||
| Both | av_tumour | dukes | STAGE | ||
| Both | av_tumour | t_best n_best m_best |
TNM STAGE | Concatenated | |
| 100kGP | cancer_analysis | histology_coded | ICD-O-3 | Removing the / character for technical reasons | |
| 100kGP | cancer_participant_tumour | morphology_snomed_ct_code morphology_icd_code |
SNOMED CT ICD-O-3 |
||
| 100kGP | cancer_participant_tumour | integrated_tnm_stage_grouping | STAGE | ||
| 100kGP | cancer_participant_tumour | ajcc_stage | STAGE | ||
| 100kGP | cancer_participant_tumour | final_figo | STAGE | ||
| 100kGP | cancer_participant_tumour | modified_dukes_stage | STAGE | ||
| 100kGP | cancer_participant_tumour | component_tnm_t component_tnm_n component_tnm_m |
TNM STAGE | Concatenated | |
| Both | cancer_registry | cancer_type cancer_behaviour |
ICD-O-3 | Concatenated | |
| 100kGP | mhsds_care_activity | codeobs obsschemeinuse |
SNOMED | if obsschemeinuse = 3 | |
| NHS-GMS | observation | normalised_hpo_id | HPO | filter value_code = present | |
| 100kGP | rare_diseases_participant_phenotype | hpo_id | HPO | filter hpo_present = true | |
| Both | sact | morphology | ICD-O-3 | ||
| Both | sact | sact_stage_at_start | STAGE | ||
| NHS-GMS | tumour | tumour_type, presentation | - | date from tumour_diagnosis_day, tumour_diagnosis_month, tumour_diagnosis_year | |
| NHS-GMS | tumour_morphology | morphology | SNOMED CT | date from parent tumour table | |
| NHS-GMS | tumour_topography | actual_body_site | SNOMED CT | date from parent tumour table | |
| NHS-GMS | tumour_topography | primary_body_site | SNOMED CT | date from parent tumour table | |
| Observation Code Description | Both | av_tumour | histology_coded_desc | ICD-O-3 | |
| Body Site Code | Both | av_tumour | site_coded | ICD-O-3 | if coding_system_desc starts with "ICD-O-3" |
| 100kGP | cancer_participant_tumour | topography_snomed_ct_code topography_snomed_code, topography_snomed_version topography_icd_code |
SNOMED CT SNOMED CT ICD-O-3 |
||
| Body Site Description | Both | av_tumour | site_coded_desc | ICD-O-3 |
Procedure¶
| Participant Explorer | Source Dataset | Source Table | Source Column(s) | Code System | Notes |
|---|---|---|---|---|---|
| Procedure Code | Both | av_treatment | eventcode | NCRAS | |
| Both | opcs4_code | OPCS-4 | |||
| Both | radiocode | NCRAS | |||
| Both | imagingcode | NCRAS | |||
| Both | imagingsite | OPCS-4 | |||
| 100kGP | cancer_invest_imaging | imaging_code_snomed_ct_code | SNOMED CT | ||
| 100kGP | cancer_surgery | primary_procedure | OPCS-4 | Ignore '.' | |
| 100kGP | rare_diseases_imaging | procedure_other_snomed_ct | SNOMED CT | ||
| Both | ecds | treatment_code_1 - treatment_code_12 | SNOMED CT | ||
| Both | hes_apc | opertn_01-24 | OPCS-4 | Z-chapter codes mapped to body site and grouped with preceding non-Z-chapter code | |
| Both | hes_op | opertn_01-24 | OPCS-4 | Z-chapter codes mapped to body site and grouped with preceding non-Z-chapter code | |
| Both | rtds | primaryprocedureopcs | OPCS-4 | ||
| Both | sact | opcs_procurement_code opcs_delivery_code |
OPCS-4 | If 3 digits: prefix with "X" Uppercase ignore "N/A" |
|
| 100kGP | did | did_snomedct_code | SNOMED CT | ||
| Procedure Code Description | Both | av_treatment | eventdesc | NCRAS | |
| Both | radiodesc | NCRAS | |||
| Both | imagingdesc | NCRAS | |||
| Body Site Code | 100kGP | cancer_invest_imaging | anatomical_site | OPCS-4 | Split comma-separated values into multiple codes for the same procedure |
| Both | hes_apc | opertn_01-24 | OPCS-4 | Z-chapter codes mapped to body site and grouped with preceding non-Z-chapter code | |
| Both | hes_op | opertn_01-24 | OPCS-4 | Z-chapter codes mapped to body site and grouped with preceding non-Z-chapter code | |
| Both | rtds | rttreatmentanatomicalsite | OPCS-4 | ||
| 100kGP | did | ic_sub_syscomp_id ic_sub_sys_id ic_system_id ic_sub_region_id ic_region_id |
SNOMED CT | Only using the most specific system code and the most specific region code. I.e., when both a region_id and sub_region_id are present, only include the sub_region_id in the body site coding. |
Medication administration¶
| Participant Explorer | Source Dataset | Source Table | Source Column(s) | Code System | Notes |
|---|---|---|---|---|---|
| Code | Both | sact | drug_group | SACT Drug Group | Non-printable characters removed and converted to title-case |
Encounter¶
| Participant Explorer | Source Dataset | Source Tables | Source Column | Notes |
|---|---|---|---|---|
| Encounter Date | Both | av_treatment | eventdate | |
| Encounter Type | Both | "National Cancer Registration" | ||
| Encounter Date | Both | av_tumour | diagnosisdatebest | |
| Encounter Type | Both | "National Cancer Registration" | ||
| Encounter Date | Both | participant | registration_date date_of_consent |
registration_date if available; otherwise date_of_consent Also used for recruited diseases |
| Encounter Type | Both | "Genomics England" | ||
| Encounter Date | 100kGP | cancer_participant_tumour | diagnosis_date | |
| Encounter Type | 100kGP | "Genomics England" | ||
| Encounter Date | 100kGP | cancer_invest_sample_pathology | event_date | |
| Encounter Type | 100kGP | "Genomics England" | ||
| Encounter Date | 100kGP | cancer_invest_imaging | imaging_date | |
| Encounter Type | 100kGP | "Genomics England" | ||
| Encounter Date | 100kGP | cancer_invest_sample_pathology | event_date | |
| Encounter Type | 100kGP | "Genomics England" | ||
| Encounter Date | 100kGP | cancer_surgery | procedure_date | |
| Encounter Type | 100kGP | "Genomics England" | ||
| Encounter Date | Both | cancer_analysis | tumour_clinical_sample_time | |
| Encounter Type | Both | "Genomics England" | ||
| Encounter Date | Both | cancer_registry | event_date | |
| Encounter Type | Both | "National Cancer Registration" | ||
| Encounter Date | 100kGP | rare_diseases_participant_phenotype | phenotype_report_date | |
| Encounter Type | 100kGP | "Genomics England" | ||
| Encounter Date | 100kGP | rare_diseases_imaging | date | |
| Encounter Type | 100kGP | "Genomics England" | ||
| Encounter Date | Both | ecds | arrival_date arrival_time |
|
| Encounter Date | Both | departure_date departure_time |
||
| Encounter Type | Both | "Emergency" | ||
| Encounter Date | Both | hes_apc | admidate | |
| Encounter Type | Both | "Inpatient" | ||
| Encounter Type | Both | hes_op | "Outpatient" | |
| Encounter Date | Both | apptdate | ||
| Encounter Type | 100kGP | did | "Diagnostic Imaging" | |
| Encounter Date | 100kGP | did_date3 | ||
| Encounter Date | 100kGP | mhmd_v4_event mhldds_event |
mhd_eventdate | |
| Encounter Type | 100kGP | "Mental Health Services" | ||
| Encounter Date | 100kGP | mhsds_medical_history_previous_diagnosis mhsds_primary_diagnosis mhsds_secondary_diagnosis |
diagdate | |
| Encounter Type | 100kGP | "Mental Health Services" | ||
| Encounter Date | 100kGP | mhsds_provisional_diagnosis | provdiagdate | |
| Encounter Type | 100kGP | "Mental Health Services" | ||
| Encounter Date | 100kGP | mhsds_indirect_activity | indirectactdate | |
| Encounter Type | 100kGP | "Mental Health Services" | ||
| Encounter Date | 100kGP | mhsds_care_activity | carecontdate | |
| Encounter Type | 100kGP | "Mental Health Services" | ||
| Encounter Date | Both | mortality | event_date | |
| Encounter Type | Both | "Office of National Statistics" | ||
| Encounter Type | Both | rtds | "Radiotherapy" | |
| Encounter Date | Both | decisiontotreatdate | For diagnosis codes | |
| Encounter Type | Both | rtds | "Radiotherapy" | |
| Encounter Date | Both | proceduredate | ||
| Encounter Date | Both | sact | date_decision_to_treat | For diagnosis, morphology and staging codes |
| Encounter Type | Both | "Chemotherapy" | ||
| Encounter Date | Both | sact | administration_date | |
| Encounter Type | Both | "Chemotherapy" | ||
| Encounter Date | NHS-GMS | observation | observation_effective_from | |
| Encounter Type | NHS-GMS | "Genomics England" | ||
| Encounter Date | NHS-GMS | tumour tumour_morphology tumour_topography |
tumour date fields (see notes) | tumour_diagnosis_day/tumour_diagnosis_month/tumour_diagnosis_year. If day not present then 01 is used. If month not present, data point is not used on the timeline. |
| Encounter Type | NHS-GMS | "Genomics England" |
-
ICD-10 codes are normalised so that they match the ICD-10 reference data. This includes the removal of any non-numeric characters other than the first character, and inserting a dot at the fourth position (for example, a coded value of "E149D" is normalised to "E14.9"). The original values can be obtained via the "Source Code" column. ↩↩↩↩↩↩↩↩↩↩↩↩↩↩↩