LabKey DOCTYPE error¶
Question
I tried loading in a LabKey table, however I ran into a lexical <!DOCTYPE html> error (invalid char in JSON text). Can I resolve this?
Answer
Luckily, the answer is most often yes! In fact, the <!DOCTYPE html> error is one of the most common errors that we receive from our users related to LabKey. Several aspects may cause this, however, the examples below are the most likely the root causes of this error. If the issue persists after you ruled out these two issues, please contact the service-desk so we can investigate.
1) folderPath issue (R users) or container_path/project_name issue (Python users)¶
The first fix consists in simply checking the path to the data programme is correct in your API call.
For R Labkey users, this is an issue with the
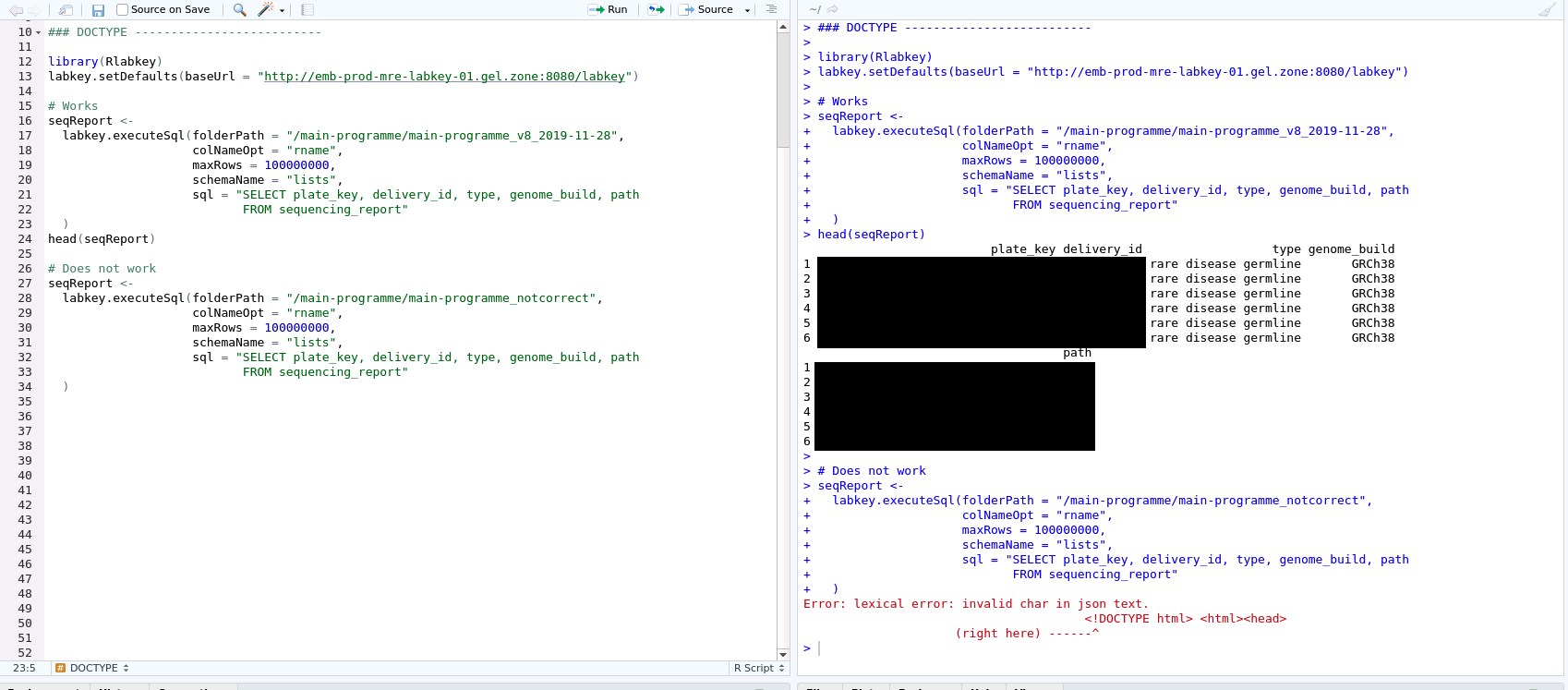
For Python users, the situation is similar: simply make sure that the second argument you pass to the "labkey.utils.create_server_context" function is spelled out correctly. This parameter is called "container_path" in the function specification, and it is usually a variable called "project_name" in our documentation. A typical example of correct spelling is this line, which you will find in our example scripts:
project_name = "/main-programme/main-programme_v19_2024-10-31" # The data we want to access. This will change depending on your study
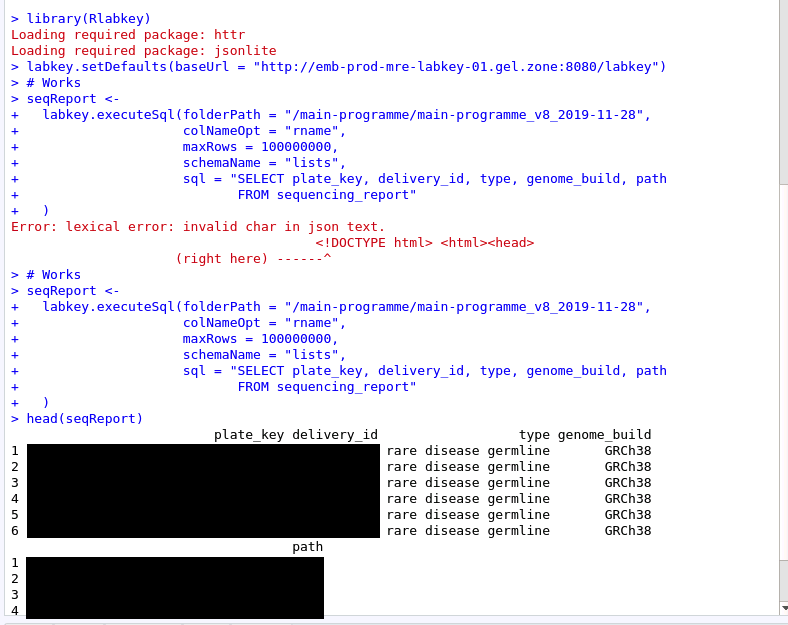
2) .netrc setup (related: recent password change)¶
The second fix relates to the .netrc file, which needs to be setup prior to using the LabKey API. This file needs to be present and to contain the correct information, including your up-to-date username and password, as explained here.
This is shown here in an example (this uses R, but the situation is similar if you use Python).
Below you can see a correct query, with the .netrc file. After I corrected my password, I resubmitted the code without actually modifying it. You will see that the output is correct, and the LabKey table has been loaded.
If you changed your password in the middle of an Rstudio session, it may be possible that you have to restart your Rstudio to recognise this change.
Last updated
This page was last updated on the 11 Feb 2020.