Creating "lollipops" diagrams in the RE¶
Question
I have a few variants of interest inside the RE, and I want to create a lollipops diagram of my gene of interest including those variants - how can I do that inside the RE?
Answer
Programs like lollipops provide this functionality, but usually require an internet connection to work.
However, note that:
- part of this task requires internet connection, in order to get the gene/protein structure information from UniProt and Pfam (this cannot be done inside the RE), but only uses public data...
- ...while part of this task uses participant data (i.e., data that cannot get out of RE), and does not require an internet connection.
Therefore, the suggestion is to "cheat" and split the task into two parts: one to be done outside the RE (e.g. on your local computer with an internet connection), and one to be done inside the RE using the lollipops program.
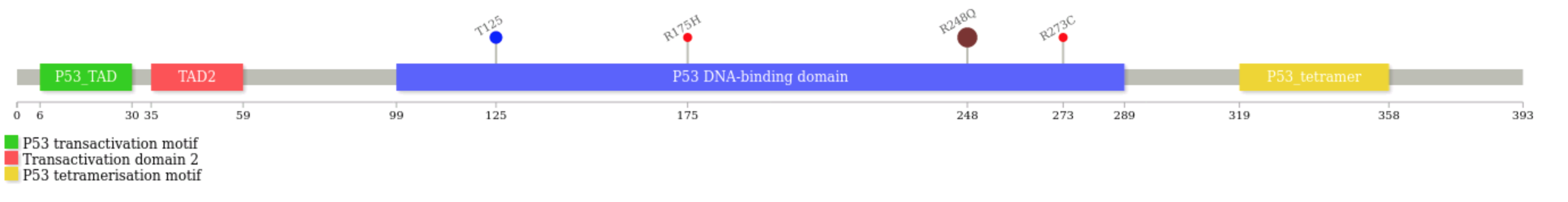
Say you have a few variants for gene TP53 and want to get your lollipops diagram:
-
Look up gene TP53 human on your machine and translate to UniProt using https://www.uniprot.org/uniprot, e.g.
(look at gene names, find TP53 human => UniProt AC is P04637)
-
Get the JSON for P04637 from the Pfam web service and copy it:
https://pfam.xfam.org/protein/P04637/graphic
(note, copy only the JSON item, i.e. do NOT copy the initial and closing square brackets, only copy everything else in between)
-
Import such text into the RE using the copy/paste (or Airlock for bulk uploads) and save it as text file
lollipops_P04637.json -
In the RE terminal, run:
module load lollipops/1.5.2 lollipops -U P04637 -l lollipops_P04637.json -legend -labels R248Q#[[email protected]](/cdn-cgi/l/email-protection) R273C R175H [[email protected]](/cdn-cgi/l/email-protection)(those are the variants at the end of the command line, in this case they are simply dummy values with no connection to Genomics England participants data - note, do not change the order of the arguments i.e. start with the "
-U" and "-l" arguments) -
Ignore the warning about fonts, and enjoy your diagram
P53_HUMAN.svg(the default output file name is.svg)
Last updated
This page was last updated on the 02 Apr 2020.